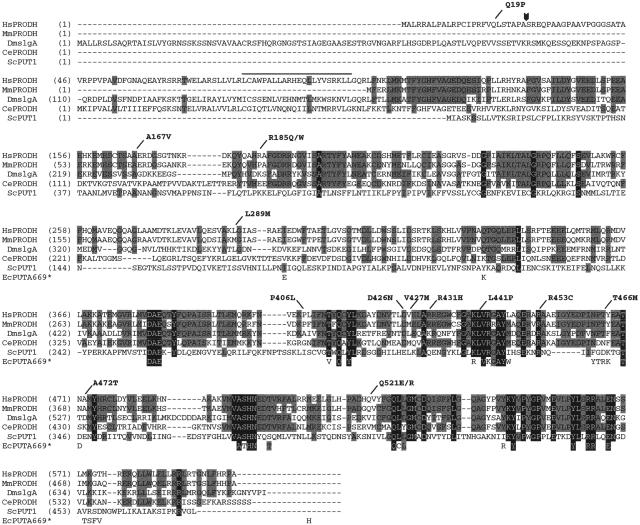
Figure 3.
Alignment of the predicted PRODH amino acid sequences from various eukaryotic species. Residues conserved across three or four of the sequences are indicated by a gray background, those conserved in all five eukaryotic examples by a black background. Also shown are the 40 residues of E. coli PutA669 that are within 5 Å of substrate and/or cofactor (Lee et al. 2003). These are spaced as they are in PutA669, but, for the sake of clarity, we have not shown the intervening residues. The shading scheme for the E. coli residues follows the convention for the eukaryotic sequences. The locations of the human missense mutations studied in this article are indicated at the top of the figure. The arrowhead indicates the predicted cleavage site for the mitochondrial targeting sequence. The overline indicates a possible leucine zipper motif. Hs=Homo sapiens, Mm=Mus musculus, Dm=Drosophila melanogaster, Ce=Caenorhabditis elegans, Sc=Saccharomyces cerevisiae, and Ec=E. coli.