Abstract
The identification of pathways that underlie common disease has been greatly impacted by the study of rare families that segregate single genes with large effect. Intracranial aneurysm is a common neurological problem; the rupture of these aneurysms constitutes a frequently catastrophic neurologic event. The pathogenesis of these aneurysms is largely unknown, although genetic and environmental factors are believed to play a role. Previous genomewide studies in affected relative pairs have suggested linkage to several loci, but underlying genes have not been identified. We have identified a large kindred that segregates nonsyndromic intracranial aneurysm as a dominant trait with high penetrance. Genomewide analysis of linkage was performed using a two-stage approach: an analysis of ∼10,000 single-nucleotide polymorphisms in the 6 living affected subjects, followed by the genotyping of simple tandem repeats across resulting candidate intervals in all 23 kindred members. Analysis revealed significant linkage to a single locus, with a LOD score of 4.2 at 1p34.3-p36.13 under a dominant model with high penetrance. These findings identify a Mendelian form of intracranial aneurysm and map the location of the underlying disease locus.
Intracranial aneurysms (IAs [MIM %105800 and MIM %608542]) represent a major public health problem. The incidence of subarachnoid hemorrhage (SAH) due to aneurysm rupture is 6 in 100,000, with ∼28,000 aneurysmal ruptures per year; it is estimated that up to 2.3% of the general population have undetected aneurysms (Juvela 2002b). The consequences of SAH are catastrophic, with approximately half of IA ruptures resulting in immediate death. Those individuals who survive the initial hemorrhage experience a 40% mortality rate during the first month, and only 25% of those who live past the first month recover completely (King 1997).
Given the poor prognosis of SAH due to aneurysm rupture, surgical or endovascular intervention prior to rupture is considered to be of paramount importance. There is, however, no practical means for reliable early diagnosis, except through screening studies of high-risk individuals with tests such as magnetic resonance angiography (MRA) or computerized tomography angiography (CTA). Moreover, there currently are no widely accepted guidelines to identify these high-risk individuals (Wermer et al. 2003).
Neither the conditions that lead to aneurysm formation nor the aneurysm ruptures are well understood. Recent studies suggest that both environmental and genetic factors contribute to the pathogenesis of IA. Risk factors for the formation, growth, and rupture of IA include hypertension, atherosclerosis, diabetes, and vascular anatomical differences (Juvela 2002a; Ohkuma et al. 2003). In addition, social factors such as smoking and diet have also been suggested to play a role in the disease (Juvela 2002b; Anderson et al. 2004).
Several lines of evidence demonstrate that genetic factors play an important role in IA pathogenesis. First, a number of genetic diseases, such as adult polycystic kidney disease (MIM #173900) (Chapman et al. 1992), Marfan syndrome (MIM #154700) (ter Berg et al. 1986), glucocorticoid remediable aldosteronism (MIM #103900) (Litchfield et al. 1998), and Ehlers-Danlos syndrome type IV (MIM #130050) (de Paepe et al. 1988), appear to increase the risk of IA formation. Second, familial recurrence of nonsyndromic IA has been well described (Fox and Ko 1980; Morooka and Waga 1983; Maroun et al. 1986; Elshunnar and Whittle 1990). Indeed, there is a three- to fivefold increase in risk for first-degree relatives of affected individuals, compared with the general population (Stehbens 1998; Ronkainen et al. 1999).
Among familial IAs, the varied pattern of recurrence has typically suggested complex causation. Several genomewide linkage studies that used affected sibling and/or relative pairs have identified various loci throughout the human genome that link to IA (Onda et al. 2001; Olson et al. 2002; Farnham et al. 2004; van der Voet et al. 2004). The results of these studies, however, have been inconsistent, and no specific disease-related genes have yet been identified. In addition, candidate-gene studies have not yet shown a robust association with IA (Onda et al. 2001; Hofer et al. 2003; Farnham et al. 2004; van der Voet et al. 2004).
In the context of substantial locus heterogeneity, the power of affected sibling pair studies or affected relative pair studies is markedly diminished (Risch and Merikangas 1996). Alternatively, the identification of rare Mendelian forms of disease can lead to the identification of genes and pathways that might play a key role in the pathogenesis of common, as well as rare, forms of the disease (Lifton et al. 2001). A limitation to this approach is that it is difficult to completely characterize and collect extended kindreds. In the present study, we have investigated what we believe is the largest-yet-reported kindred with IA; genomewide analysis of linkage provides significant evidence that the disease in this family is attributable to inheritance of a single locus at 1p34.3-p36.13.
Since 1994, we have screened >3,000 patients with IAs and identified 142 kindreds with two or more affected members. The family investigated in this study (IA 20) is the largest in our database and has been described elsewhere (Fox and Ko 1980). At that time, there were six members with proven IA, all in generation II (fig. 1). The pedigree has since been extended and further characterized. In total, there are now 10 documented subjects with IAs, 1 subject with distinctive multiple intracranial vessel occlusions and extensive collateral vessel formation of unknown etiology (subject III-3), and 1 subject with abdominal aortic aneurysm (AAA) at a young age (age 32 years) (individual II-5); this latter trait is sometimes associated with IA (Cannon Albright et al. 2003). For the purpose of linkage analysis, the patients with documented IAs and the patient with multiple intracranial vascular occlusions were classified as “affected,” and the patient with AAA was prospectively classified as “phenotype unknown.” There are also 12 unaffected descendents of subject I-2. Of these, 8 were asymptomatic at >30 years of age and had negative screening results for magnetic resonance imaging, MRA, CTA, and/or catheter angiogram (individuals II-3, II-6, II-12, II-14, III-1, III-2, III-8, and IV-2); 3 offspring of unaffected subjects were asymptomatic at >30 years of age and did not have screening studies (individuals III-5, III-9, and III-10); and 1 individual was asymptomatic at <30 years of age, without screening studies (individual IV-1). For linkage studies, this latter subject was classified as “phenotype unknown,” and the others were classified as “unaffected.”
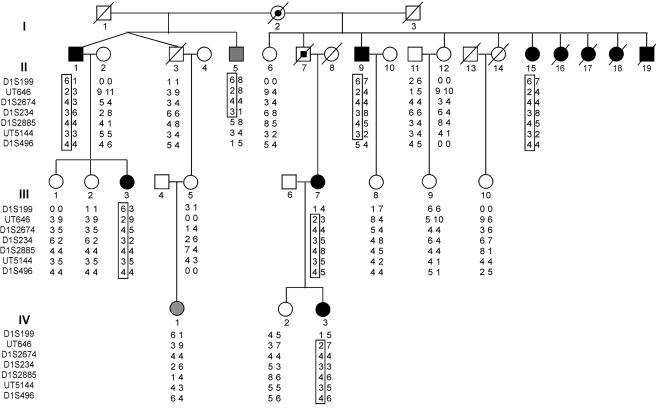
Figure 1.
IA 20 kindred. Affected and unaffected individuals are shown as blackened and unblackened symbols, respectively. Obligate carriers are shown as partially blackened symbols. Individuals II-5 and IV-1 were assigned an affection status of “unknown” prior to linkage analysis and are shown as gray symbols. The genotypes of STR marker loci that span 14 cM at 1p34.3-p36.13 are shown, and segments of the haplotype linked to the disease phenotype are enclosed in a box. An allele designated “0” indicates a failed reaction.
The clinical features of the affected members are presented in table 1. Age at diagnosis of IA ranged from 21 years to 53 years by MRA or CTA prior to SAH (n=7) and from 29 years to 57 years for patients who presented with SAH (n=4). There are scant risk factors for IA among kindred members; specifically, there is a history of hypertension in only one individual, and, although smoking was prevalent among both affected and unaffected family members, there was no significant difference between the two groups (8 of 10 affected individuals were smokers, and 10 of 12 unaffected individuals were smokers). Specifically, there is no history of polycystic kidney disease (no history of end-stage renal disease and no serum creatinine level >1.5 mg/dl), no history of Marfan syndrome (no history of aortic dissection, ectopia lentis, etc.), and no history of Ehlers-Danlos syndrome (no history of hypermobile joints, hyperextensible skin, or easy scarring). Finally, in neither the affected-only genomewide linkage analysis nor the affected-plus-unaffected analysis (see below) was there evidence of linkage to known loci for any of these syndromes. Members of both sexes are affected, the trait is present in consecutive generations, all affected members are the offspring of either known or suspected IA cases, and approximately half the offspring of such subjects have IA (fig. 1). These findings are consistent with autosomal dominant transmission of IA with high penetrance.
Table 1.
Clinical Features of Affected Members of Kindred IA 20[Note]
| ID | Aneurysm Location | Age at Diagnosis(years) |
| II-1 | ACoA | 38 |
| II-7 | ACA, left MCA | 53 |
| II-9 | ACoA | 40 |
| II-15 | Left MCA | 29 |
| II-16 | Left MCA (SAH) | 32 |
| II-17 | OphtA (SAH) | 57 |
| II-18 | Right MCA, ACoA (SAH) | 32 |
| II-19 | Left ICA (SAH) | 29 |
| III-3 | Bilateral MCA occlusion | 30 |
| III-7 | Left MCA | 36 |
| IV-3 | Basilar, right MCA × 2 | 21 |
Note.— ACA = anterior cerebral; ACoA = anterior communicating; ICA = internal carotid; MCA = middle cerebral; OphtA = ophthalmic arteries.
Blood samples were collected from all available family members after the obtainment of their informed consent; the study protocol was approved by the Yale Human Investigations Committee. Genomic DNA was prepared as described elsewhere (Bell et al. 1981).
We used a two-stage design in linkage analysis (Elston et al. 1996). We first genotyped all available affected individuals (n=6) by use of Affymetrix 10K GeneChips. The SNPs genotyped on these chips provide an estimated information content equivalent to a microsatellite screen density of 1 marker/1–2.5 cM (Kruglyak 1997). SNP genotypes were obtained by following the Affymetrix protocol for the GeneChip Mapping 10K Xba Array. Briefly, genomic DNA was digested with XbaI; adapters were ligated to the product and an adapter-specific primer was used to amplify the product by PCR. The products were purified, fragmented, and labeled with biotin-ddnTP. Biotin-labeled DNA fragments were hybridized to the Mapping 10K Array chip. After hybridization, arrays were washed, stained, and scanned. Affymetrix MicroArray Suite 5.0 software was used to obtain raw microarray feature intensities, which were processed using the Affymetrix Genotyping Tools software package to derive SNP genotypes.
An average of 9,468 genotypes was scored per subject (SNP call-rate range, 91%–97%). To analyze the GeneChip data for linkage, we created a UNIX-based program (Chunky) that parses the data sheet into individual files per chromosome in linkage format. The information captured includes chromosome number, SNP markers, map distances, genotype calls, and allele frequencies.
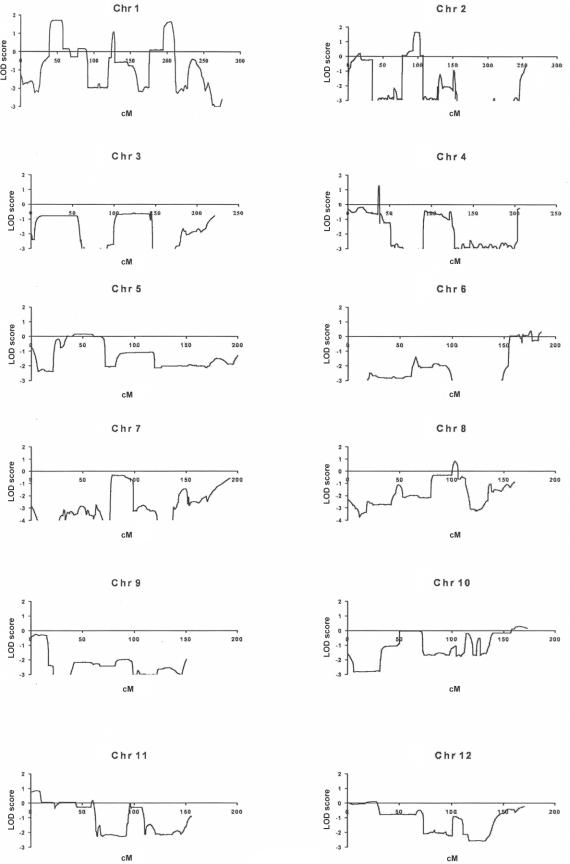
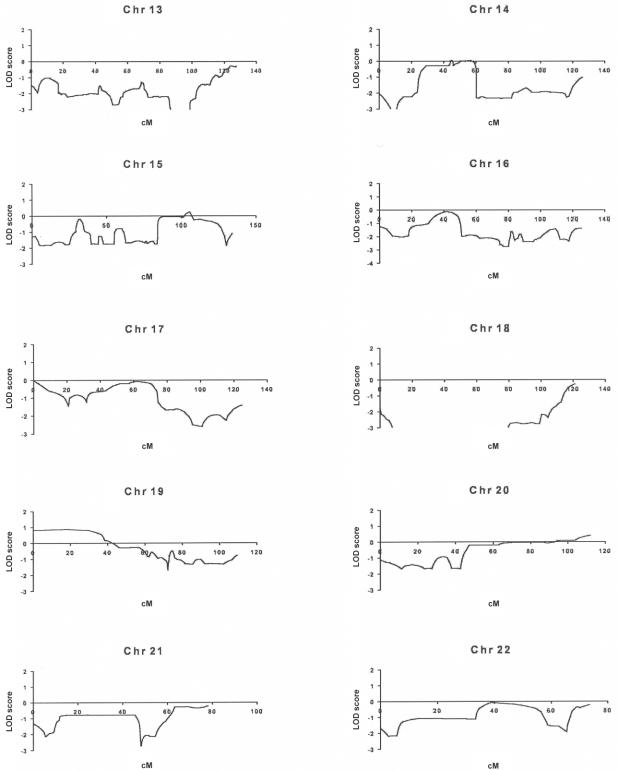
For the multipoint analysis of linkage, we specified the disease locus as autosomal dominant, with penetrance that varied from 70% to 99%, a mutant disease-gene frequency of 0.001, and a phenocopy rate of 0.001. SNP allele-frequency data for the white population, as supplied by Affymetrix, were used for the analysis of linkage, which was performed using the Allegro program (deCODE). This analysis identified three intervals with LOD scores near the theoretical maximum of 1.8 (1p34.3-p36.13, 1q31-q41, and 2p11-p14), with LOD scores of <0 for nearly all of the remainder of the genome (fig. 2). The LOD scores were confirmed using the GENEHUNTER program. In an additional analysis, we specified the trait locus as X-linked dominant, with otherwise similar estimates of the trait locus; no interval on the X chromosome achieved a LOD score >−0.2. Additional genotyping with GeneChip of four unaffected individuals yielded only these same three intervals with LOD scores of ⩾1.0. Changing the phenocopy rate had small effects on the LOD scores and did not identify additional candidate intervals.
Figure 2.
Analysis of linkage in the IA 20 kindred from GeneChip data of affected individuals. Linkage graphs for all chromosomes are shown; the X-axis corresponds to genetic distance (in cM), and the Y-axis shows LOD scores.
Using data from the University of California–Santa Cruz (UCSC) Genome Browser (May 2004) (UCSC Genome Bioinformatics Web site), we identified and genotyped from five to nine highly polymorphic di- and tetranucleotide microsatellite markers across each of the three candidate intervals in all available kindred members (n=23). Genotyping for microsatellite analysis was performed by PCR, with detection of fluorescent products on an ABI 3700 sequencer (Applied Biosystems) equipped with GeneScan and Genotyper software (Applied Biosystems). The results were analyzed using the Simwalk program (we specified marker heterozygosities of 75% and the same autosomal dominant model of the trait locus used above, with penetrance of 70%–99%).
Our analysis diminished the evidence of linkage to 1q31-q41 and 2p11-p14 (table 2). In contrast, it demonstrated that all affected members inherit the same haplotype at 1p34.3-p36.13, whereas this haplotype was transmitted to none of the unaffected members (fig. 1). Parametric linkage analysis (with 99% penetrance specified) yielded a maximum LOD score of 4.2 at 1p34.3-p36.13 (table 2 and fig. 3); changing estimates of marker-allele frequencies had negligible effects on the LOD score. The likelihood of linkage to 1p34.3-p36.13 was nearly 1,000-fold more likely than the next-most-likely interval at 1q31-q41 (table 2).
Table 2.
Maximum LOD Scores for Linkage of STRs and IA[Note]
|
Maximum LOD Score,for Penetrance of |
|||
| Interval | 70% | 90% | 99% |
| 1p34-1p36 | 3.4 | 3.9 | 4.2 |
| 1q31-1q41 | 1.3 | −.1 | −5.6 |
| 2p11-2p14 | −.3 | −2.3 | −6.6 |
Note.— Maximum LOD scores are reported for 1p34-1p36, 1q31-1q41, and 2p11-2p14 for STR markers in all family members, with varying estimates of penetrance.
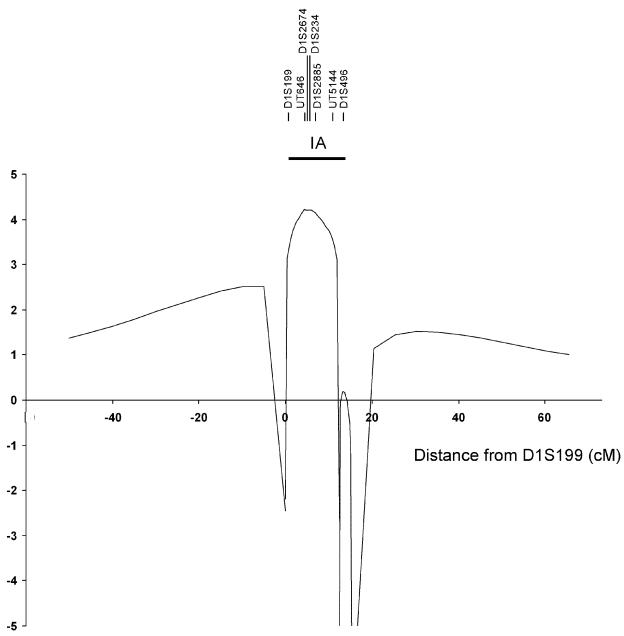
Figure 3.
Analysis of linkage with STR markers on 1p34.3-p36.13. An IA gene is localized to a 12.5-cM region between markers D1S199 and D1S496, with a maximum LOD score of 4.2. Multipoint analysis of linkage, for comparison of segregation of IA with marker loci, was performed. The location of marker loci used is indicated at the top of the diagram. The Y-axis shows LOD scores. The horizontal bar indicates the extent of the LOD-1 interval.
The LOD score peak occurs at UT646; the LOD-1 interval is flanked by loci D1S199 and D1S496 (fig. 3), which define a 12.5-cM interval that corresponds to a 15-Mb segment (from 19.3 million bp to 34.9 million bp). This is the same interval defined by the GeneChip analysis, which indicated a LOD-1 interval flanked by rs950922 and rs514262 that corresponded to a 15.4 million–bp interval (from 21.3 million bp to 36.7 million bp on 1p34.3-p36.13).
Analysis of critical recombinants supports localization of the IA locus within the specified interval. Affected subject II-9 is recombinant at the distal border, and subject III-7 is recombinant at the proximal border (fig. 1). Nearly identical borders define the linked interval by SNP analysis.
Examination of the LOD-1 interval identified ∼240 genes. Among these, a number of genes have been identified as plausible candidate genes, including polycystic kidney disease–like 1, brain-specific angiogenesis inhibitor 2, fibronectin type III domain–containing gene, and collagen type XVI α1.
To our knowledge, the present kindred is the largest yet reported with IA, with 10 definitively affected subjects and one likely affected subject. Genomewide analysis of linkage in this kindred demonstrates complete linkage of IA to a 12.5-cM segment of chromosome 1, with evidence of linkage that substantially exceeds thresholds for significance. The phenotyping in the kindred was clear-cut; reclassification of the patient with multivessel occlusions and extensive collateral growth as “phenotype unknown” would reduce the maximum LOD score to 3.9. Moreover, the LOD score was substantially increased by inclusion of unaffected family members, which supports high penetrance of the trait locus. It is also of note that the subject with the early AAA inherited a segment of the linked haplotype, which suggests that this vascular aneurysm might be attributable to inheritance at this same locus. It would be of interest to obtain abdominal ultrasounds in kindred members to determine whether this phenotype commonly cosegregates with IAs and/or linked haplotypes. The pattern of segregation and the linkage data indicate that this family defines a new Mendelian form of IA that is transmitted as an autosomal dominant trait with high penetrance. Similar to reported cases of familial IAs, members of IA 20 presented with SAH or symptomatic findings at an earlier age than is typically found in sporadic cases (Lozano and Leblanc 1987).
These findings represent a first step in the identification of a susceptibility gene for IA. Other than young age, there are no obvious clinical features that separate IA in members of this family from typical cases in the general population. It is presently unknown whether the locus implicated in this study might play a role in other common forms of IA. In principle, it is possible that this might be a one-of-a-kind family with a rare mutation that results in a highly penetrant form of IA. It is also possible that other less-penetrant mutations in the same gene or pathway play a role in more-common forms of IA. To date, a number of studies have used linkage approaches to attempt to identify loci that contribute to IA risk (de Paepe et al. 1988; Pope et al. 1990; Kuivaniemi et al. 1993; Takenaka et al. 1999; Onda et al. 2001; Olson et al. 2002; Yoneyama et al. 2003; Farnham et al. 2004). Sib pair studies from Japanese and Finnish populations, as well as a recent study of a consanguineous Dutch family, have identified candidate intervals (Onda et al. 2001; Olson et al. 2002; Roos et al. 2004). The only intervals from such studies that meet genomewide evidence of significant linkage are 19q13.3 in the Finnish population (van der Voet et al. 2004) and 2p13 in the Dutch family (Roos et al. 2004).
The identification of the causative gene in IA 20 will shed light on the pathways that lead to disease. Whether this locus or pathway will play a role in more-common forms of disease remains to be determined. However, once genes that lead to IAs are identified, they may better define the pathophysiology and natural history of aneurysm formation and rupture. Finally, these findings may contribute to improved diagnostic and therapeutic approaches to this disease.
Acknowledgments
We are grateful to the members of the kindred studied for their invaluable contribution to this study. We thank Anita Farhi, R.N., and Andrea Chamberlain, R.N., for their help with recruitment of the family members. We also thank the Keck Affymetrix GeneChip and Genotyping Facilities at Yale for assistance with the SNP genotyping. The study was supported in part by the Yale General Clinical Research Center and by an institutional award from the Howard Hughes Medical Institute.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for IA, adult polycystic kidney disease, Marfan syndrome, glucocorticoid remediable aldosteronism, and Ehlers-Danlos syndrome type IV)
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/
References
- Anderson CS, Feigin V, Bennett D, Lin RB, Hankey G, Jamrozik K (2004) Active and passive smoking and the risk of subarachnoid hemorrhage: an international population-based case-control study. Stroke 35:633–637 [DOI] [PubMed] [Google Scholar]
- Bell GI, Karam JH, Rutter WJ (1981) Polymorphic DNA region adjacent to the 5′ end of the human insulin gene. Proc Natl Acad Sci USA 78:5759–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon Albright LA, Camp NJ, Farnham JM, MacDonald J, Abtin K, Rowe KG (2003) A genealogical assessment of heritable predisposition to aneurysms. J Neurosurg 99:637–643 [DOI] [PubMed] [Google Scholar]
- Chapman AB, Rubinstein D, Hughes R, Stears JC, Earnest MP, Johnson AM, Gabow PA, Kaehny WD (1992) Intracranial aneurysms in autosomal dominant polycystic kidney disease. N Engl J Med 327:916–920 [DOI] [PubMed] [Google Scholar]
- de Paepe A, van Landegem W, de Keyser F, de Reuck J (1988) Association of multiple intracranial aneurysms and collagen type III deficiency. Clin Neurol Neurosurg 90:53–56 [DOI] [PubMed] [Google Scholar]
- Elshunnar KS, Whittle IR (1990) Familial intracranial aneurysms: report of five families. Br J Neurosurg 4:181–186 [DOI] [PubMed] [Google Scholar]
- Elston RC, Guo X, Williams LV (1996) Two-stage global search designs for linkage analysis using pairs of affected relatives. Genet Epidemiol 13:535–558 [DOI] [PubMed] [Google Scholar]
- Farnham JM, Camp NJ, Neuhausen SL, Tsuruda J, Parker D, MacDonald J, Cannon-Albright LA (2004) Confirmation of chromosome 7q11 locus for predisposition to intracranial aneurysm. Hum Genet 114:250–255 [DOI] [PubMed] [Google Scholar]
- Fox JL, Ko JP (1980) Familial intracranial aneurysms: six cases among 13 siblings. J Neurosurg 52:501–503 [DOI] [PubMed] [Google Scholar]
- Hofer A, Hermans M, Kubassek N, Sitzer M, Funke H, Stogbauer F, Ivaskevicius V, Oldenburg J, Burtscher J, Knopp U, Schoch B, Wanke I, Hubner F, Deinsberger W, Meyer B, Boecher-Schwarz H, Poewe W, Raabe A, Steinmetz H, Auburger G (2003) Elastin polymorphism haplotype and intracranial aneurysms are not associated in central Europe. Stroke 34:1207–1211 [DOI] [PubMed] [Google Scholar]
- Juvela S (2002a) Hypertension and aneurysmal subarachnoid hemorrhage. Wien Klin Wochenschr 114:285–286 [PubMed] [Google Scholar]
- ——— (2002b) Natural history of unruptured intracranial aneurysms: risks for aneurysm formation, growth, and rupture. Acta Neurochir Suppl 82:27–30 [DOI] [PubMed] [Google Scholar]
- King JT Jr (1997) Epidemiology of aneurysmal subarachnoid hemorrhage. Neuroimaging Clin N Am 7:659–668 [PubMed] [Google Scholar]
- Kruglyak L (1997) The use of a genetic map of biallelic markers in linkage studies. Nat Genet 17:21–24 [DOI] [PubMed] [Google Scholar]
- Kuivaniemi H, Prockop DJ, Wu Y, Madhatheri SL, Kleinert C, Earley JJ, Jokinen A, Stolle C, Majamaa K, Myllyla VV (1993) Exclusion of mutations in the gene for type III collagen (COL3A1) as a common cause of intracranial aneurysms or cervical artery dissections: results from sequence analysis of the coding sequences of type III collagen from 55 unrelated patients. Neurology 43:2652–2658 [DOI] [PubMed] [Google Scholar]
- Lifton RP, Gharavi AG, Geller DS (2001) Molecular mechanisms of human hypertension. Cell 104:545–556 [DOI] [PubMed] [Google Scholar]
- Litchfield WR, Anderson BF, Weiss RJ, Lifton RP, Dluhy RG (1998) Intracranial aneurysm and hemorrhagic stroke in glucocorticoid-remediable aldosteronism. Hypertension 31:445–450 [DOI] [PubMed] [Google Scholar]
- Lozano AM, Leblanc R (1987) Familial intracranial aneurysms. J Neurosurg 66:522–528 [DOI] [PubMed] [Google Scholar]
- Maroun FB, Murray GP, Jacob JC, Mangan MA, Faridi M (1986) Familial intracranial aneurysms: report of three families. Surg Neurol 25:85–88 [DOI] [PubMed] [Google Scholar]
- Morooka Y, Waga S (1983) Familial intracranial aneurysms: report of four families. Surg Neurol 19:260–262 [DOI] [PubMed] [Google Scholar]
- Ohkuma H, Tabata H, Suzuki S, Islam MS (2003) Risk factors for aneurysmal subarachnoid hemorrhage in Aomori, Japan. Stroke 34:96–100 [DOI] [PubMed] [Google Scholar]
- Olson JM, Vongpunsawad S, Kuivaniemi H, Ronkainen A, Hernesniemi J, Ryynanen M, Kim L-L, Tromp G (2002) Search for intracranial aneurysm susceptibility gene(s) using Finnish families. BMC Med Genet 3:7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda H, Kasuya H, Yoneyama T, Takakura K, Hori T, Takeda J, Nakajima T, Inoue I (2001) Genomewide-linkage and haplotype-association studies map intracranial aneurysm to chromosome 7q11. Am J Hum Genet 69:804–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope FM, Limburg M, Schievink WI (1990) Familial cerebral aneurysms and type III collagen deficiency. J Neurosurg 72:156–158 [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Ronkainen A, Niskanen M, Piironen R, Hernesniemi J (1999) Familial subarachnoid hemorrhage: outcome study. Stroke 30:1099–1102 [DOI] [PubMed] [Google Scholar]
- Roos YB, Pals G, Struycken PM, Rinkel GJ, Limburg M, Pronk JC, van den Berg JS, Luijten JA, Pearson PL, Vermeulen M, Westerveld A (2004) Genome-wide linkage in a large Dutch consanguineous family maps a locus for intracranial aneurysms to chromosome 2p13. Stroke 35:2276–2281 [DOI] [PubMed] [Google Scholar]
- Stehbens WE (1998) Familial intracranial aneurysms: an autopsy study. Neurosurgery 43:1258–1259 [DOI] [PubMed] [Google Scholar]
- Takenaka K, Sakai H, Yamakawa H, Yoshimura S, Kumagai M, Yamakawa H, Nakashima S, Nozawa Y, Sakai N (1999) Polymorphism of the endoglin gene in patients with intracranial saccular aneurysms. J Neurosurg 90:935–938 [DOI] [PubMed] [Google Scholar]
- ter Berg HW, Bijlsma JB, Veiga Pires JA, Ludwig JW, van der Heiden C, Tulleken CA, Willemse J (1986) Familial association of intracranial aneurysms and multiple congenital anomalies. Arch Neurol 43:30–33 [DOI] [PubMed] [Google Scholar]
- van der Voet M, Olson JM, Kuivaniemi H, Dudek DM, Skunca M, Ronkainen A, Niemela M, Jaaskelainen J, Hernesniemi J, Helin K, Leinonen E, Biswas M, Tromp G (2004) Intracranial aneurysms in Finnish families: confirmation of linkage and refinement of the interval to chromosome 19q13.3. Am J Hum Genet 74:564–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wermer MJ, Rinkel GJ, van Gijn J (2003) Repeated screening for intracranial aneurysms in familial subarachnoid hemorrhage. Stroke 34:2788–2791 [DOI] [PubMed] [Google Scholar]
- Yoneyama T, Kasuya H, Onda H, Akagawa H, Jinnai N, Nakajima T, Hori T, Inoue I (2003) Association of positional and functional candidate genes FGF1, FBN2, and LOX on 5q31 with intracranial aneurysm. J Hum Genet 48:309–314 [DOI] [PubMed] [Google Scholar]