Abstract
Recent advancements in RNA therapeutics highlight the critical need for precision gene delivery systems that target specific organs and cells. Lipid nanoparticles (LNPs) have emerged as key vectors in delivering mRNA and siRNA, offering protection against enzymatic degradation, enabling targeted delivery and cellular uptake, and facilitating RNA cargo release into the cytosol. This review discusses the development and optimization of organ- and cell-specific LNPs, focusing on their design, mechanisms of action, and therapeutic applications. We explore innovations such as DNA/RNA barcoding, which facilitates high-throughput screening and precise adjustments in formulations. We address major challenges, including improving endosomal escape, minimizing off-target effects, and enhancing delivery efficiencies. Notable clinical trials and recent FDA approvals illustrate the practical applications and future potential of LNP-based RNA therapies. Our findings suggest that while considerable progress has been made, continued research is essential to resolve existing limitations and bridge the gap between preclinical and clinical evaluation of the safety and efficacy of RNA therapeutics. This review highlights the dynamic progress in LNP research. It outlines a roadmap for future advancements in RNA-based precision medicine.
Keywords: Lipid carriers, RNA therapies, DNA barcoding, Site-specific, Personalized medicine, Gene delivery
1. The rise of siRNA and mRNA therapeutics and the need for precision delivery
1.1. Lipid nanoparticle-enabled mRNA therapeutics
Ribonucleic acid (RNA)-based therapeutics such as small interfering RNA (siRNA) and messenger RNA (mRNA) are on the rapid rise in clinical development for genetic medicines. The most well-known clinical applications are perhaps the two mRNA-based COVID-19 vaccines from Moderna (Elasomeran/Spikevax®) and Pfizer/BioNTech (Tozinameran/Comirnaty®), both using ionizable lipid nanoparticles (LNPs) as non-viral gene delivery systems for formulating the mRNAs [1,2]. LNPs play a critical role in shielding the RNAs from ribonuclease (RNase)-mediated degradation and, more significantly, facilitating the escape of RNA from endolysosomal degradation to enter the cytosol — a process known as endosomal escape — upon the LNPs’ cellular uptake via endocytosis [3,4]. As depicted in Fig. 1 (left panel), LNP-formulated mRNAs are essential for the in vivo translation of both endogenous and exogenous peptides/proteins. This delivery method enables the direct production of biological therapeutics, such as vaccination antigens [2], therapeutic proteins, e.g., erythropoietin [5], antibody therapeutics [6,7], novel cellular receptors, e.g., the chimeric antigen-receptors (CARs) for cellular immunotherapies [8], and agents for genome editing, specifically those encoding CRISPR-Cas9 mRNA and single-guide RNA (sgRNA) [9–11]. The incorporation of a replicase sequence upstream of the therapeutic mRNA transforms it into self-amplifying RNA (saRNA), enhancing the translation of therapeutic proteins [12]. Beyond the clinically approved mRNA COVID-19 vaccines, both Moderna and BioNTech are developing over forty mRNA therapeutics for a range of applications, from Zika virus vaccines [13], personalized cancer vaccines [14], tolerogenic autoimmune/allergy vaccines [15,16], and various protein therapeutics, [17] e.g., Human erythropoietin (hEPO) [18]. The recent FDA approval of the first CRISPR-Cas9/sgRNA-based therapy (exagamglogene autotemcel/Casgevy™, delivered ex vivo via electroporation) further highlighted the potential of mRNA in encoding CRISPR-Cas for gene editing in various preclinical studies [19–22]. Significant milestones were achieved with two-phase I trials of lung targeting CRISPR-mRNA-LNPs dosed in 2024 for treating cystic fibrosis (RCT2100, NCT06237335) and primary ciliary dyskinesia (RCT1100, NCT05737485), highlighting the rapidly expanding use of the LNP-enabled gene editing [23].
Fig. 1. Mechanism of Action of mRNA and siRNA therapeutics.
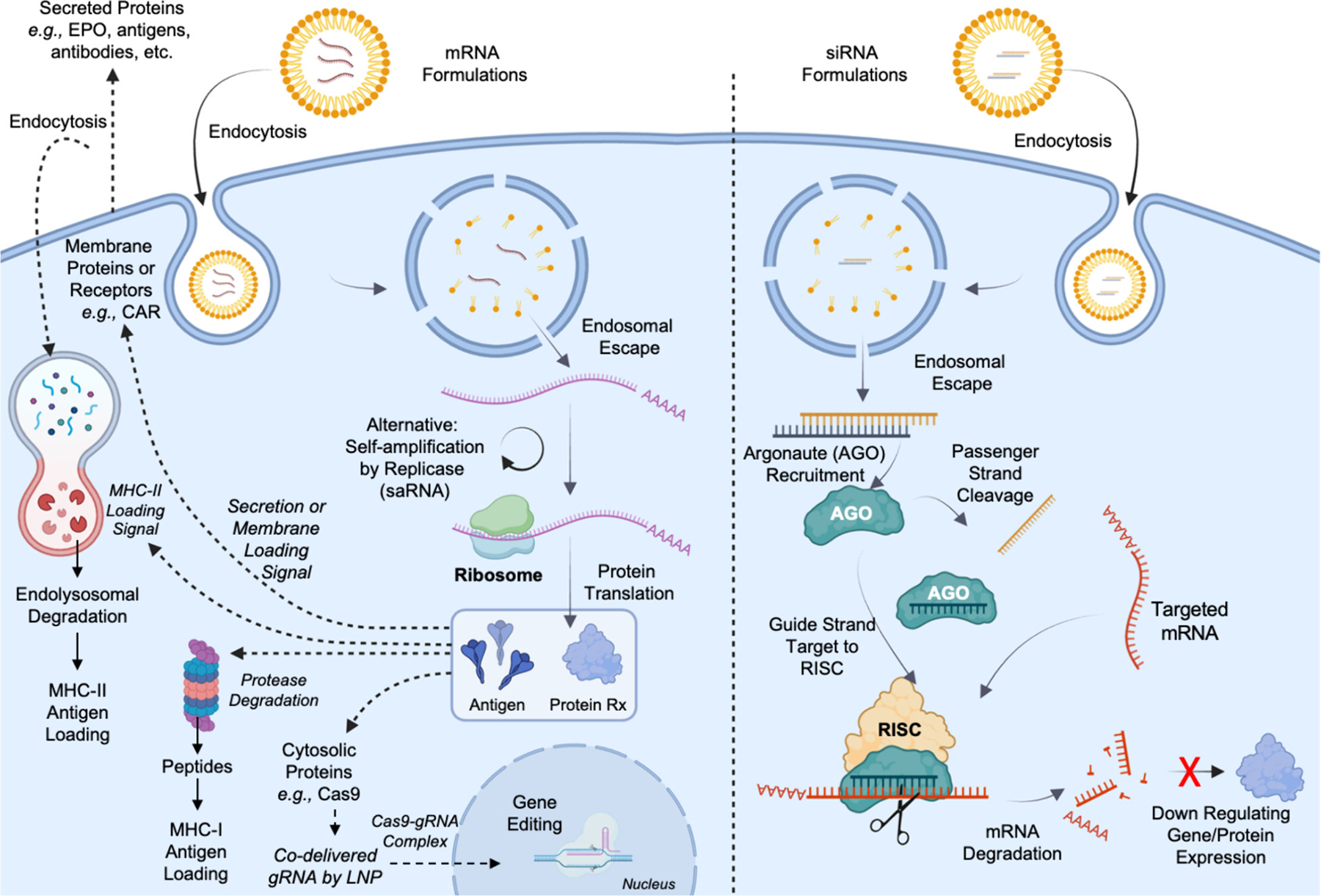
mRNA and siRNA therapeutics both aim to alter protein functions by enabling protein translation or inhibiting protein translation. When formulated in lipid nanoparticles, both enter cells via endocytosis, which makes the “endosomal escape” of RNA-LNPs the primary biological barrier in their bioavailability. By carefully designing RNA sequences, mRNA serves as multifunctional platforms to produce intracellular proteins or antigens, membrane-bounded proteins or receptors, and secreted proteins that could also be recycled and endocytosed by the cell (left panel). On the contrary, siRNA enabled sequence-specific degradation of mRNA through the recruitment of RNA-induced Silencing Complex (RISC), leading to down-regulation of protein expression (right panel), which could serve as a stand-alone therapy or as emergency brakes for mRNA therapies to counteract any unintended consequences of mRNA therapy by silencing the same mRNA (This figure is created with BioRender.com).
1.2. siRNA therapeutics enabled by N-Acetylgalactosamine (GalNAc)-conjugates and lipid nanoparticle formulations
siRNA molecules, typically 20–25 nucleotides per strand and forming a double-stranded structure, play a crucial role in the RNA interference (RNAi) pathway. These double-stranded RNAs modulate gene expression by recruiting the RNA-induced Silencing Complex (RISC), which results in the targeted degradation of mRNA and the suppression of gene and protein expression (see Fig. 1, right panel) [24]. Initiation of this process involves the Argonaute protein (AGO) family, which facilitates the removal of the passenger (sense) strand of the siRNA [25], thus allowing the guide (antisense) strand to direct the RISC to the complementary mRNA sequence. This interaction leads to mRNA cleavage and degradation, silencing gene function [26,27]. Endogenously, siRNAs contribute to the regulation of gene expression and genomic stability. Exogenously, they have been synthesized for targeted gene knockdown in mammalian cells since 2001 [28]. The clinical trajectory of siRNA therapeutics began with the FDA approval of Patisiran (Onpattro®) in 2018 for the treatment of hereditary transthyretin amyloidosis (hATTR) [29]. Following this, four additional siRNA drugs have been authorized for hATTR (Vutrisiran/ Amvuttra®), acute hepatic porphyria (Givosiran/ Givlaari®), primary hyperoxaluria type 1 (Lumasiran/ Oxlumo®), and to lower low-density lipoprotein cholesterol (Inclisiran/ Leqvio®) [30–33].
1.3. The rising need for precision delivery of siRNA and mRNA therapeutics
siRNA therapeutics exhibit distinct dose-dependent pharmacodynamics and require periodic administration to sustain their efficacy [34], similar to small molecule drugs, except that siRNA therapeutics have significantly longer dosing intervals ranging from every 3 weeks to every 6 months [35]. The enduring effect of siRNA is unique and distinct from traditional sustained-release/long-acting dosage forms known for small molecular drugs. This necessitates a greater need for precision in RNA therapeutic delivery as the off-target effect will also become long-lasting [36,37]. In contrast, mRNA therapeutics often result in transient protein production [38]. Therefore, the most effective approach to harnessing their therapeutic potential is often through leveraging the immune responses, as seen with mRNA vaccines. A two-dose regimen can induce a robust and sustained immune response. To ensure strength and safety, careful design and precision delivery of mRNA therapeutics are essential. This maximizes efficacy and minimizes the risk of auto-immunity and other off-target effects [39]. In the context of CRISPR/Cas9 gene editing, the consequences of the off-target effects could be significant [40,41]. Precision RNA delivery is critical when utilizing LNPs for delivering Cas9-encoding mRNA and sgRNA. Achieving organ-specific and cell-specific delivery for genome editing remains one of the biggest challenges for ensuring safety and efficacy [41,42]. As the field of RNA therapeutics burgeons, the success of these modalities hinges on the precision of delivery systems to minimize off-target effects [43]. While siRNA and mRNA therapies offer distinct pharmacological advantages and challenges, their full potential can only be capitalized with precision delivery.
Following the rapid development of both RNA therapeutics and lipid nanoparticles (LNPs), recent reviews have addressed various aspects of this multidisciplinary field, such as organ-specificity [44], non-liver targeting LNPs [45], spleen-targeted LNPs [46], mRNA-LNPs design [47,48], and clinical trials of siRNA therapeutics [49], etc. This review aims to serve as a one-stop shop by holistically examining the history and advancements in liposomal RNA delivery systems. We focus on state-of-the-art organ-specific and cell-specific RNA delivery strategies using LNPs and discuss the latest applications of DNA/RNA barcoding technologies for enhancing the targeting-specificity of LNPs. Additionally, we summarize current clinical trials of mRNA and siRNA therapeutics to highlight these innovative RNA therapies’ translational potential and future directions.
2. A 60-year journey from liposomes to RNA-delivering ionizable lipid nanoparticles
Liposomal membraned formulations such as liposomes and LNPs are clinically approved therapeutic carriers capable of delivering small molecules and biologics such as proteins, peptides, and nucleic acids [50–52]. Liposomes and LNPs are self-assembled in aqueous buffers but are structurally different from each other. Liposomes typically consist of one to multiple lipid bilayers for entrapping large amounts of aqueous buffer inside the lipid bilayer membrane, often referred to as the aqueous core of the liposomes. LNPs’ core predominately comprises lipids and hydrophilic small water pockets formed by ionizable lipids and nucleic acids in their inner core [53], shelled by a lipid mono-layer [54] or lipid bilayer membrane [55]. As demonstrated by the COVID-19 mRNA vaccine, LNPs have demonstrated their transformative potential for enabling RNA therapeutics. Finding the path for enabling this journey is a collective effort built over 60 years of research on lipid chemistry and formulations, as summarized in Fig. 2 (see Table S1 for indexed references).
Fig. 2. A 60-year journey from liposomes to ionizable lipid nanoparticles for RNA delivery.
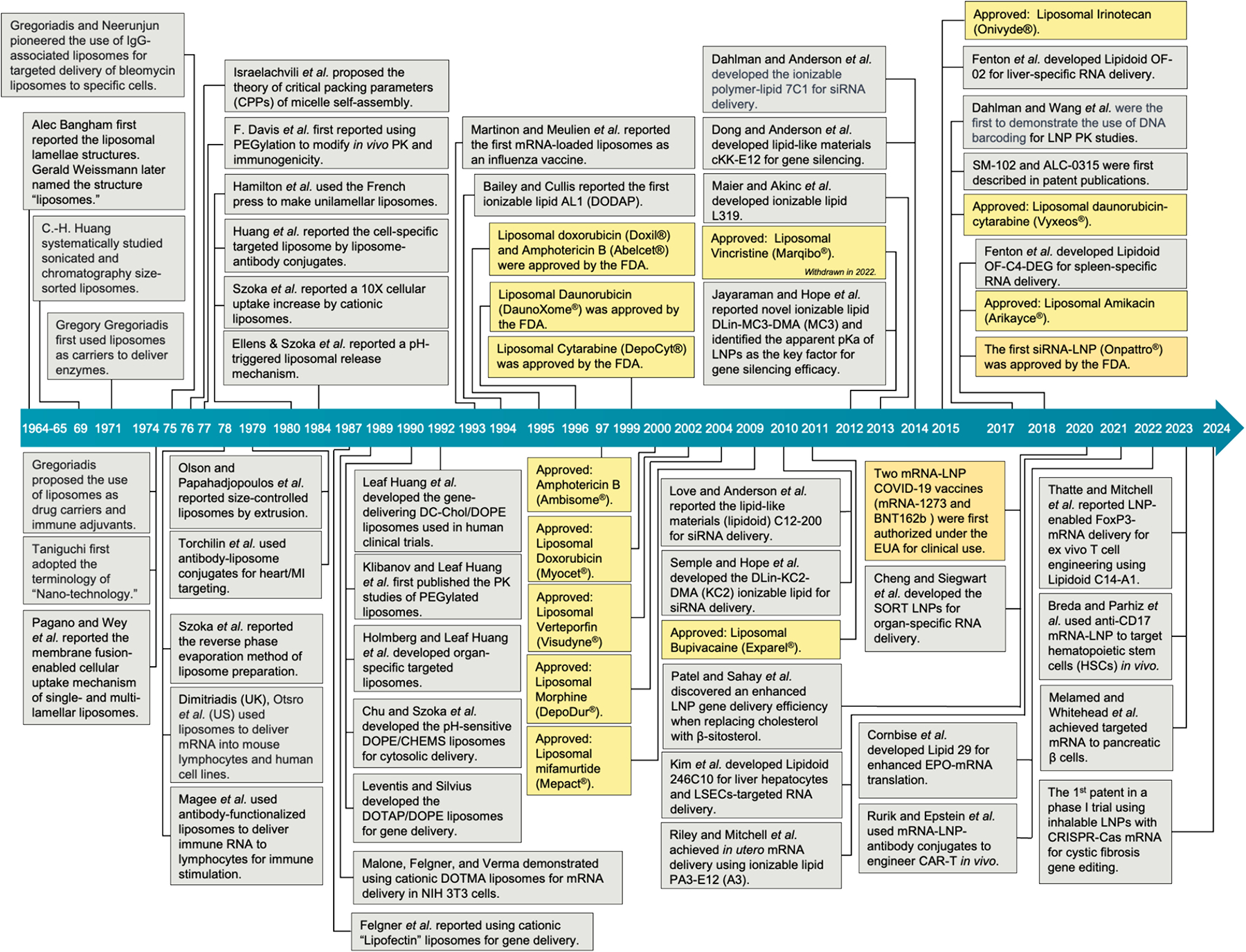
This retrospective timeline summarizes the development of liposomes and lipid nanoparticle formulation, with selected milestones highlighting key discoveries for drug and gene delivery. Full itemized information is summarized in Table S1.
2.1. Early discoveries of liposomes for therapeutic delivery
The journey into the realm of the minuscule, which would eventually lead to the development of RNA-delivering liposomal structures, was inspired by Richard Feynman’s visionary 1959 talk, “There’s Plenty of Room at the Bottom.” This lecture sparked a wave of research focused on the scientific exploration at the microscopic scales and the advancement of sophisticated microscopy techniques. Coinciding with this surge was Alec Bangham’s seminal microscopy discovery of liposomal lamellar structures in 1964, initially described as “swollen phospholipids.” By controlling the sizes and properties of the liposomal lamellar structures using sonication and chromatography [56], these phospholipid vesicles, resembling cellular membranes, have since evolved into highly biocompatible carriers capable of encapsulating diverse therapeutic agents as delivery systems, first proposed and demonstrated by Gregory Gregoriadis in the early 70s for in vivo enzyme delivery [57–59]. Coinciding with the burst of nanotechnology [60], liposomes soon evolved from basic biomembrane/vesicles to versatile therapeutic carriers due to their ability to encapsulate biological molecules [58,59,61]. In 1978, one year after Ostro et al. incorporated high molecular weight RNA into liposomes, Dimitriadis (UK) and Ostro et al. (US) independently reported their successful use of liposomal formulations for delivering mRNA for inducing protein translation in mouse lymphocytes and human cell lines [62,63]. This officially opened the era of developing gene-delivering liposomal formulations. During that time, liposomal formulations typically comprised egg- or soy-derived charge-neutral phosphatidylcholine (PC) lipids with a blend of varying amounts of cholesterol. Encapsulating hydrophilic RNAs into PC-based liposomes has one major challenge, which is the low encapsulation efficiency when using simple lipid film hydration methods (i.e., making liposomes by dissolving hydrophilic cargo such as RNAs in aqueous buffers then vortexed it with dried lipid film to foster self-assembly of multi-lamellar liposomes). In the same year, F. Szoka, Jr. and D. Papahadjopoulos reported the “reverse phase evaporation method” that largely improves the encapsulation efficiency of hydrophilic molecules. This is achieved by forming reverse-micelle emulsion from the lipids using an immiscible buffer and solvent pair, followed by solvent evaporation to form large unilamellar liposomes [64]. The lipid film hydration method and reverse phase evaporation have been widely used in studying RNA delivery since then.
2.2. Cationic liposomes for gene delivery
Building on early works that studied cellular interactions of positively charged liposomes, which showed higher cell association compared to negatively charged liposomes [65], cationic lipids, e.g., N-[1-(2,3-dioleyloxy)propyl]-N,N,N-trimethylammonium chloride (DOT MA), were first reported for use in mRNA delivering liposomes in the late 80s [66,67]. Cationic liposomes complex nucleic acids via charge interactions, forming mixtures known as “lipoplexes.” At that time, lipoplexes were used as a much-improved gene transfection method, compared to the cationic polymer-based transfection method using diethylaminoethyl-dextran (DEAE-Dextran) and the calcium phosphate co-precipitation methods developed in the 60s’ and 70s’ [68,69]. The advancement of cationic liposomes for gene delivery finally entered human clinical trials with leading formulation using cationic cholesterol derivatives with cationic lipid blend, i.e., DC-Chol/DOPE [70–73]. Parallel to cationic liposomes is the development of pH-sensitive/responsive liposomal formulations by Szoka et al. with compositions like pH-responsive cholesterylhemisuccinate (CHEMS) and cationic dioleoylphosphatidylethanolamine (DOPE), which significantly improved the cytosolic delivery of non-pH-sensitive formulation such as the mixture of CHEMS and neutrally charged dioleoylphosphatidylcholine (DOPC) [74]. This is achieved by incorporating pH-sensitive lipids capable of changing their self-assembly properties, thus destabilizing the lipid bilayer at lower pH values, resembling the acidifying process of the endosome to lysosome trafficking when liposomes are endocytosed [75]. Escaping endolysosomal trafficking is recognized as a critical biological barrier for the efficacy of liposome-encapsulated mRNA and siRNA.
2.3. The critical packing parameters theory of lipid self-assembly
Liposomes and LNPs are both made of self-assembled lipid structures, yet they are structurally different, driven by their functional need to encapsulate different therapeutic molecules. Despite the differences, liposomes and LNPs share nearly identical engineering principles, such as the critical packing parameters (CPPs) theory of lipids/micelles self-assembly. The concept of CPPs was first proposed by Israelachvili et al. in 1976, which provides a generalized self-assembly theory linking thermodynamics, molecular interactions free energies, and geometry [76]. The CPP is represented by this equation: Ns = Vc / ae * Lc, where Vc = the volume of the hydrophobic chain, i.e., the volumetric space occupied by the hydrophobic part of the molecule, which is typically averse to water. The Lc = length of the hydrophobic chain, usually given in nanometers (nm). The ae = the area per molecule at the hydrophilichydrophobic interface also called the effective headgroup area. This is the area occupied by the hydrophilic (water-attracting) part of the molecule at the interface where the molecule meets water. It’s an important factor in determining how the molecules pack together. Amphiphilic surfactants and soap molecules typically have an Ns ≪ 1/3, representing their smaller hydrophobic tails compared to the hydrophilic headgroups, leading to a conical-shaped molecule favoring the formation of spherical micelles. A 1/3 ≤ Ns < 1/2 typically forms cylindrical micelles. A flexible bilayer is typically formed when 1/2 ≤ Ns < 1, and the planer bilayer is formed when Ns values ≈ 1. When Ns > 1, the lipids will favor the formation of inverse micelles or other structures where the hydrophilic headgroups are internalized. Assembling lipid building blocks with different CPPs into liposomes or LNPs is like packing differently shaped objects (representing the lipid molecules) into boxes (representing the space in water). Depending on the shape of the objects, nature arranges them differently to fit (self-assemble) most efficiently. Similarly, the shape of the lipid molecules (influenced by their CPP) determines how they’ll pack together in water, leading to different assembled structures. The ability of lipids to assemble into varying structures depending on the geometry of the lipid components is known as “lipid polymorphism.”
2.4. The development of ionizable lipids and lipid nanoparticles for gene delivery
Observations on cationic liposomal formulations for gene delivery have led to extensive lipid polymorphism studies that ultimately give birth to the first ionizable cationic lipids 1,2-dioleoyl-3-dimethylammonium propane (DODAP, also known as AL1) in 1994 by Bailey and Cullis [77]. DODAP liposomes have an apparent pKa of 6.58, which forms cationic liposomes at pH 4 when co-formulated with helper supporting lipids such as phosphatidylcholine (PC), dioleoylphosphatidylethanolamine (DOPE), and cholesterol. However, fusion of lipid membranes was observed when pH raised from 4.0 to 7.5 due to the formation of one particular lipid polymorph, i.e., the H₂ Phase (Hexagonal Phase). The H₂ phase refers to a specific arrangement in which the lipid molecules organize themselves into cylindrical tubes that pack together in a hexagonal pattern, destabilizing lipid bilayers. This observations on pH-dependent lipid polymorph of ionizable lipids as the pH decreases, from L2 (inverse micelle), H2 (hexagonal), and Q2 (cubic) to lamellar structures, have led to the creative use of ionizable lipids such as D-Lin-MD3-DMA (MC3) or RNA delivery via forming LNPs [78]. This enabled the first FDA approval of LNP-formulated siRNA Patisiran (Onpattro; Alnylam) in 2018 [79–82]. Similar lipid molecular designs and pH-dependent self-assembly principles were adapted for ionizable lipids ALC-0315 (BioNTech/Pfizer) and SM-102 (Moderna) for delivering the COVID-19 mRNA vaccine [78]. As shown in Fig. 3a, gene-delivering ionizable lipids such as MC-3 and SM-102 have a much larger Vc and a much smaller head group, yielding a CPP >1, conducive to forming pH-responsive, non-lamellar structures like inverse micelles, with internal ionizable heads and external hydrophobic tails. [68,240]. In contrast, membrane lipids such as DSPC with a CPP approximately equal to 1 result in a bilayer structure due to its cylindrical shape, with hydrophilic heads at the water interface and hydrophobic tails tucked away (Fig. 3b). At lower pH levels, the ionizable amines on the SM-102 headgroups become cationic, facilitating electrostatic complexation with anionic nucleic acids. This interaction ensures the nucleic acids are securely encapsulated within the interior of the ionizable inverse micelles, with the hydrophobic tails of SM-102 oriented outward. However, such a hydrophobic exterior is inherently unstable in aqueous environments. To overcome this, additional membrane-forming structural supporting lipids are required for the formation of stable LNPs. This external lipid membrane layer provides a hydrophilic interface with the aqueous medium and enhances colloidal stabilities, leading to the final LNP formulations used in the clinics (Fig. 3c).
Fig. 3. Demonstration of the Critical Packing Parameter (CPP) and its application in designing ionizable lipids for effective mRNA delivery in lipid nanoparticles.
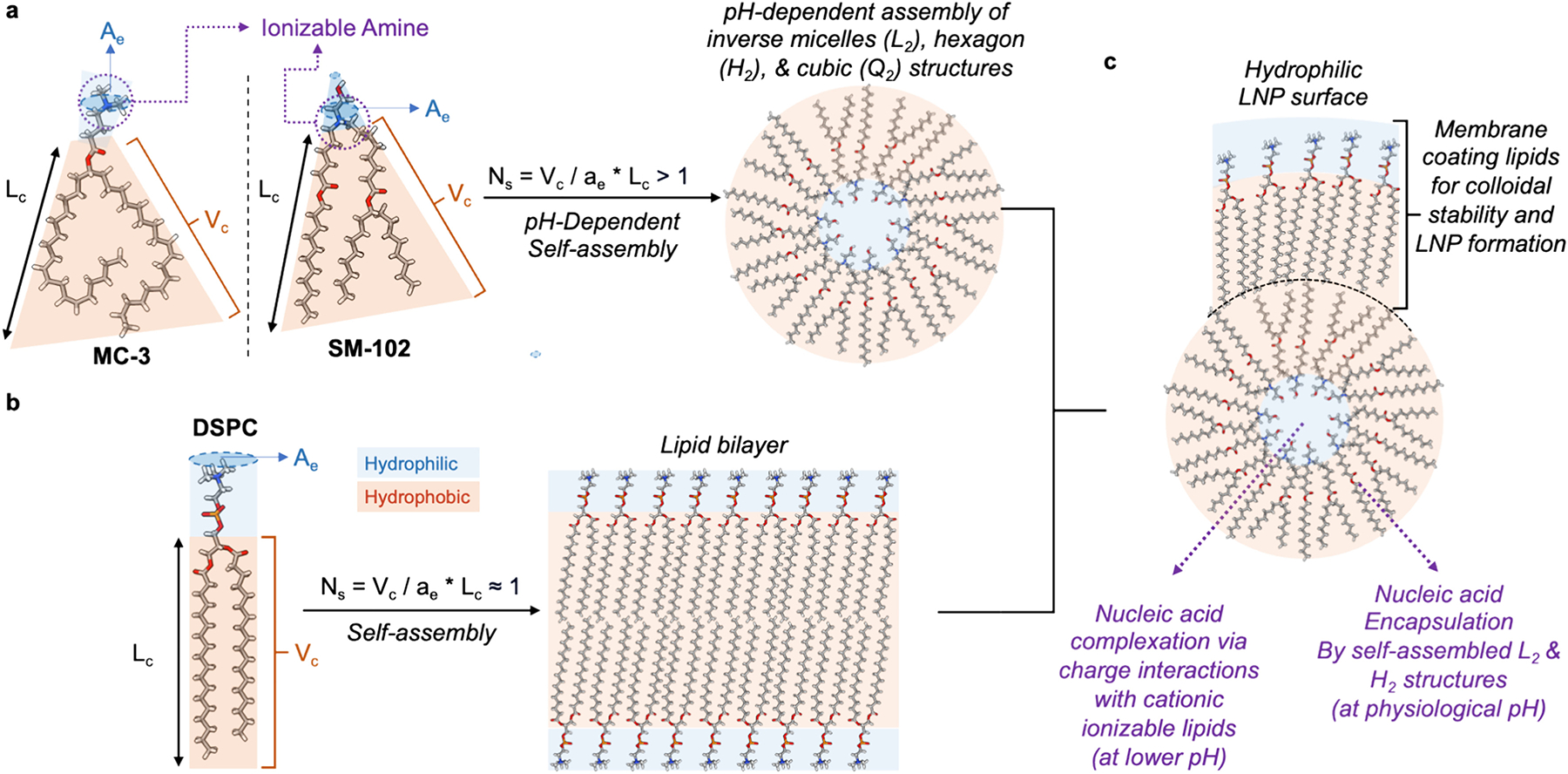
CPP is a determinant of lipid molecular arrangement in aqueous environments, defined by the equation Ns = Vc / ae * Lc, where Vc = the volume of the hydrophobic chain, Lc = length of the hydrophobic chain, ae = the effective surface area of the hydrophilic head. The most likely self-assembled structures of lipid molecules in water may differ depending on their corresponding CPP. (a) Gene-delivering ionizable lipids such as MC-3 and SM-102 have a much larger Vc and a much smaller head group, yielding a CPP >1, conducive to forming pH-responsive, non-lamellar structures like inverse micelles, with internal ionizable heads and external hydrophobic tails. [78,268]. (b) In contrast, membrane lipids such as DSPC with a CPP approximately equal to 1 result in a bilayer structure due to its cylindrical shape, with hydrophilic heads at the water interface and hydrophobic tails tucked away. (c) At lower pH levels, the ionizable amines on the SM-102 headgroups become cationic, facilitating electrostatic complexation with anionic nucleic acids. This interaction ensures the nucleic acids are securely encapsulated within the interior, with the hydrophobic tails of SM-102 oriented outward. However, such a hydrophobic exterior is inherently unstable in aqueous environments. To overcome this, additional membrane-forming lipids are incorporated into the nanoparticle formulation. These lipids assemble into a stabilizing hydrophilic monolayer around the exterior, providing a compatible interface with the aqueous medium for enhanced colloidal stabilities, which is essential for the practical application of the lipid nanoparticles for mRNA delivery.
3. Overviews of lipid components in liposomes and lipid nanoparticle formulations
The rapidly evolving field of LNPs for RNA delivery has seen an explosion of new cationic and ionizable lipids, making it challenging to remain abreast of developments without a foundational understanding of the lipids involved. Fig. 4 categorizes the lipids utilized in liposome and LNP formulations into seven primary groups: saturated and unsaturated fatty acids (Fig. 4a, section 3.1); phospholipids, e.g. phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphate (PA) (Fig. 4b, section 3.2); polyethylene glycol conjugated (PEGylated) lipids (Fig. 4c, section 3.3); cationic lipids and helper lipids (Fig. 4d, section 3.4); cholesterol and sterol-derivatives (Fig. 4e, section 3.5); ionizable inverse micellar lipids (Fig. 4f, section 3.6); and ionizable lipidoids (lipid-like multi-tail molecules, Fig. 4g, section 3.7). This simplified classification provides rough guidance in the vast chemical landscape of lipids used in lipid-formulated delivery systems. In the forthcoming sections, we aim to provide an account for these lipid categories, offering insights into the cutting-edge advancements in lipid-facilitated RNA delivery.
Fig. 4.

Lipids for formulating RNA delivering ionizable lipid nanoparticles.
3.1. Saturated and unsaturated fatty acids
Fatty acids (Fig. 4a) are the basic building blocks of the hydrophobic lipid tails and major factors for determining critical packing parameters: the length of the lipid chain (Lc) and the volume of the hydrophobic lipid chain (Vc). To drive the critical packing parameter towards inverse micelles, unsaturated fatty acids with one to five unsaturated bonds are often used to increase the bulkiness of the lipid tails when synthesizing gene-delivering ionizable lipids.
3.2. Phospholipids
PCs such as DPPC (dipalmitoyl phosphatidylcholine) and DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine) are fundamental components in assembling lipid bilayers (Fig. 4b). These bilayers, which are biomimetic mirrors of cell membranes and nuclear envelopes, excel in encapsulating nucleic acids and serving as selective barriers for molecular transport. The intrinsic properties of these bilayers underpin the design of liposomes and LNPs, providing the necessary encapsulation and protection for therapeutic delivery. The bilayer’s behavior and functionality hinge on the physicochemical characteristics of the membrane structural lipids. The lipid tails’ length and saturation level govern the bilayer’s fluidity and phase/melt transition temperature (Tm). Lipid bilayers below Tm are packed in a more organized gel state with lower membrane mobility and permeability. Bilayer above the Tm has more lipid mobility, leading to less order packing and higher membrane permeability. Long saturated tails confer increased stability and a higher phase transition temperature, resulting in more rigid bilayers compared to short saturated lipid tails. Conversely, the incorporation of unsaturated fatty acids like oleic and linoleic acid disorganized the lipid bilayer, leading to significantly reduced Tm, enlarging the hydrophobic volume of the tails, altering the critical packing parameter, and decreasing the bilayer stability [83,84]. Lipid headgroups also influence the Tm, which generally follows the trend of PC < PS (phosphatidylserine) < PE (phosphoethanolamine) < PA (phosphate) [85]. In addition to the melt transition temperature, the hexagonal phase transition (Th) of PE is used to describe the transition between the lamellar bilayer and the inverse hexagonal phase, which destabilizes the bilayer (bad for liposome formation) and favors inverse hexagonal assembly (good for encapsulating RNAs in LNPs, to be stabilized by lipid mono-layer, as shown in Fig. 3c). Similar to the Tm, the presence of unsaturated lipid tails also significantly reduces the Th for PE [78,86,87].
3.3. PEGylated lipids
PEGylation, i.e., chemically conjugating polyethylene glycol on biomolecules, was first reported by Frank Davis in 1978 as a strategy to reduce immunogenicity while increasing blood circulation time [88]. The discovery of the enhanced permeation and retention (EPR) effect of macromolecular in solid tumors in 1986 [89] has led to great interest in developing PEGylated lipids (e.g., DSPE-PEG2kDa, Fig. 4c), which have since been tested in liposomes and found to significantly increase blood circulation time, first reported in the literature by Leaf Huang et al. in 1990 [90], which ultimately led the first approval of liposomal drug, the PEGylated liposomal doxorubicin in 1995 [91]. Through prolonged blood circulation, PEGylated liposomes are able to take advantage of the EPR effect to enhance tumor drug accumulation, leading to a series of clinical approvals of liposomal chemotherapeutics, e.g., daunorubicin, cytarabine, irinotecan, etc. (Fig. 2 and Table S1), which opened the golden era of NanoMedicine and bio-nano interfacial studies [92–98,269,270]. Many more liposomal chemotherapeutics are still in development, especially for those capable of inducing cancer immunogenic cell death, e.g., mitoxantrone and oxaliplatin [50–52,99,100]. PEGylated lipids are also present in LNPs, e.g., DMC-C-PEG2kDa (Patisiran/Alnylam), DMG-mPEG2kDa (Moderna), and ALC-0159 (Pfizer/BioNTech), due to their critical roles for enhancing formulation stabilities and altering LNP’s pharmacokinetics upon systematic injections [101–103].
3.4. Cationic lipids and helper lipids
Cationic Lipids, e.g., DOTAP (1,2-dioleoyl-3-trimethylammonium-propane), DOGS, DOSPA, DOTMA, etc. (Fig. 4d), are positively charged amino lipids that are essential for the formation of complexes with negatively charged phosphodiester backbone of RNA molecules to form the “lipoplex.” Their primary function is to encapsulate the RNA and protect it from enzymatic breakdown. The cationic charges also promote cellular uptake by fostering electrostatic attractions with the cell membranes, which are generally negatively charged. Compared to DOTMA, the first cationic lipid used for RNA delivery, DOTAP changes the lipid tail conjugation from the ether (more stable) to ester (more degradable and cleavable), which is more biocompatible/biodegradable but less efficient in gene transfection in vivo [104]. DOPE (dioleoylphosphatidylethanolamine, Fig. 4b), a helper lipid, when combined with DOTMA or DOTAP, adds to the flexibility and fusogenic potential of the liposome or LNP membrane [105]. This fusion is crucial for the delivery system, as it aids in merging with the cell membrane and facilitating the release of RNA into the target cells. Liposomes and LNPs made of DOTAP/DOPE can be tailored to exhibit a range of zeta potentials, resulting in cell-type varying gene delivery efficiency in vitro [106].
3.5. Cholesterol and sterol-derivatives
Sterols (Fig. 4e), such as cholesterol, are planar, rigid, hydrophobic molecules present in abundance for stabilizing natural cell membranes [107,108]. Increasing cholesterol concentration in the bilayer inhibits lipid movements, thus decreasing membrane fluidity, making the bilayer more rigid. When used in high concentrations, such as in clinically approved liposomal formulations (nearly 40% in molar ratio), the melting phase transition temperature of the liposomes could completely disappear, leaving highly stable liposomes suitable for drug encapsulation [109]. In the context of gene delivery, cholesterol serves as a helper lipid for cationic DOTAP formulations to stabilize the lipoplexes and improve gene transfer efficiency [110]. As successful gene delivery requires the use of membrane-destabilizing lipids, cholesterol is also routinely used in LNP formulations to stabilize the formulation. Substituting cholesterol with other sterol derivatives has been shown to alter LNP biodistribution and gene delivery efficiency [111,112]. Alternatively, acidic cholesterol derivatives such as cholesteryl hemisuccinate (CHEMS) are used to formulate pH-responsive gene delivery liposomes when co-formulated with non-bilayer lipid DOPE. CHEMS is negatively charged at natural pH, which stabilizes DOPE to form liposomes. When protonated at a lower pH and losing the negative charge, the CHEMS can no longer stabilize DOPE; thus, the bilayer is destabilized, favoring the gene delivery [113]. This ionizable property of CHEMS has also been used to stabilize bilayer-embedded cationic lipid drug conjugations for tumor targeting [52]. On the other hand, Cationic derivatives of cholesterol, such as DC-Chol, could also be co-formulated with DOPE as cationic liposomes as the first cationic liposomes being tested in clinical trials in 1992 [70,71]. More recently, ionizable lipid MC3-DMA-inspired DMAPA-CHEMS and dual-cholesterol cross-linked amino-phosphate (CAP2) were able to form ionizable LNPs for siRNA and mRNA delivery [114,115]. In addition to chemically synthesized cholesterol derivatives, natural sterols such as β-sitosterol were found to enhance the endosomal escape of LNPs, facilitating RNA delivery [111,116]. Replacing cholesterol with bile acid was found to alter the biodistribution of mRNA-LNPs and reduce liver distribution [112].
3.6. Ionizable inverse micellular lipids
The earliest developed ionizable lipid is the 1,2-dioleoyl-3-dimethylammonium-propane (DODAP, also known as AL1, Fig. 4f) [77]. DODAP is structurally similar to the cationic gene delivering lipid 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) with just one difference in the head group. DODAP has a tertiary amine head (ionizable) compared to DOTAP’s quaternary amine head group (always cationic). This ionizable head enables pH-dependent lipid polymorph, as mentioned in section 2.4, favoring hexagonal/inverse micellular assembly with a critical packing parameter >1. The ionizable head group of DODAP (ester tail) and DODMA (ether tail) results in less toxicity compared to cationic cousins DOTAP (ester tail) and DOTMA (ether tail) [117]. Increasing the numbers of cis-unsaturated bonds in the DOTMA tails generates D-Lin-DMA (di-linoleyloxy tails, two cis-double bonds/tail) and D-Len-DMA (di-linolenyloxy tails, three cis-double bonds/tail). D-Lin-DMA was found to be the most fusogenic and the most efficient one for siRNA delivery and gene knock-down, followed by D-Len-DMA, with DOTMA being the least efficient of the three lipids [118]. Continuously optimizing the head group chemistry landed a pKa value at the sweet spot of ~6.4, which led to the clinical use of DLin-MC3-DMA (MC3) [29,82,119]. Structured derivatized from MC3, lipid L319 introduced ester-conjugated unsaturated tails for enhanced biodegradability, followed by a head group-modified version ATX-001 [120]. Similarly, TCL053 kept the MC3 head group but replaced the linoleyloxy tails with three ester/degradable lipid tails, exhibiting low immunogenicity for repeated dosing in skeletal muscle [121]. Ionizable lipids with amino alcohol head groups, e.g., ALC-0315 and SM-102, showed enhanced endosomal escape with enhanced mRNA translation; both adopted cleavable ester linkages to reduce in vivo circulation half-life and live accumulation (compared to MC3) and thereby enhance biocompatibility [122,123]. Derivatizing from SM-102 to replace the hydroxyl headgroup with squaramide led to the Lipid 29 with significantly increased in vivo mRNA-protein translation for model protein hEPO [18]. Ester conjugated cleavable tail design has dominated the chemical space of ionizable inverse micellular lipids, e.g., A9 [124], IR-117-17 (A10-Lin) [22], RCB-4–8 [21], PL-1 [125], L101 [126], RM-133-3 [127], LP-01 [128,129], C24, 3-A2-7b [20], and DB-11-10-8 [130], etc. Replacing ether/ester with thioether and bio-reducible disulfide bonds has also been attempted in 93-O17S [131], 8-O14B [132], and O12B/O16B/N16B derivatives, providing alternative degradation mechanism in the ionizable lipid designs [133,134].
3.7. Ionizable lipidoids
Parallel to the development of the traditionally looking ionizable lipids, high-throughput polymer synthesis using ultra-large-scale automation-assisted combinatorial chemistry rapidly expanded the chemical space for gene delivery biomaterials, screening hundreds to thousands of structural derivatives of polycationic, pH-responsive, and biodegradable polymers in the early 2000s by Akinc, Anderson, Lynn, and Langer et al. [135–139]. This is achieved using amine and parallel synthesis compatible chemical reactions, e.g., Michael addition (reacting with alkyl-acrylate or alkyl-acrylamides), reductive amination [140], ring-opening reactions, etc. [141]. Using the parallel synthesis toolboxes validated in prior polymer works for synthesizing “lipid-like” molecules called “lipidoids.” The first lipoids library contained a molecular library with amide linkages, two or more alkyl tails, 8–12 carbons in alkyl tails, and secondary amines. A total of 1200+ lipidoids were synthesized from alkyl-acrylate, alkyl-acrylamide, and amino molecules building blocks using Michael addition by Akinc et al. in 2008 for screening siRNA-delivering lipids with the Lipoids 98N12–5 as the top candidate (Fig. 4g) [142]. For in vivo siRNA silencing, lipidoid derivatives with amide conjugated tail exhibited significantly higher siRNA gene silencing efficiency compared to lipidoids with ester-conjugated tail. This is similar to the observation from cationic lipids between DOTMA (ether tail) and DOTAP (ester tail) mentioned previously [106]. When formulated into siRNA-LNPs, over 90% of the 98N12–5 LNPs were distributed to the liver [143].
On the other hand, Love et al. reported a second-generation lipidoid library of 126 lipidoids synthesized by nucleophilic epoxide ring opening reaction. They discovered the top candidate, C12–200, which is also a polycationic lipidoid stabilized by a piperazine ring with five nondegradable amino alcohol lipid tails. siRNAs delivered in vivo by C12–200 LNPs were found to be 100 times more efficient than 98N12–5 (LNP01), which has since become benchmarking lipidoids for siRNA delivery [144], derivatizing into many structurally similar lipidoids with 4–5 non-cleavable lipid tails via amine-mediated nucleophilic epoxide ring opening for varying applications, e.g., 246-C10 (4-tails) [145], C14–4 (ether-linked, 5 tails, for CAR-T engineering) [8], C14-A1 (stretched version, 5 tails), 144 Gen1-C4E12 (4 tails with a cleavable disulfide bond), PA3-C12 (A-3 for in utero mRNA delivery) [146], C12–113 (TLR7/8 agonist) [147], OC2-K3-E10 (muscle mRNA delivery) [148], PPZ-A10 (immune cell targeting) [149], and C14-O2 (CAR-macrophages/monocytes) [150], etc. In parallel, Whitehead et al. looked into developing lipidoids with cleavable ester-conjugated tails for siRNA delivery [141]. A total of 1400+ lipidoids with alkyl-acrylates ester builder blocks for generating easter-conjugated cleavable tails. siRNAs delivered in vivo by lead lipidoid 304O13 were found to be similarly potent compared to siRNAs C12–200, yet the cleavable 304O13 exhibited much lower liver toxicity compared to the non-cleavable C12–200 at higher siRNA/LNP doses [151].
Learning from pair-wise comparisons between cationic DOTMA and DOTAP, ionizable lipids DODAP and DODMA, and lipidoids 98 N12–5 amide vs. ester tail studies, ester-conjugated lipid tails often performed more poorly for in vivo RNA delivery due to rapid degradation although being more rapidly cleared from the body makes them more biocompatible [106,143]. Lipidoids with diketopiperazine (DKP) core structures were synthesized by Dong et al., led to the discovery of cKK-E12 lipidoid with an ApoE-facilitated hepatocyte uptake mechanism (similar to 246-C10 [145]), leading to more efficient in vivo siRNA-gene silencing compared to C12–200 [152]. Fenton et al. further derivatized cKK-E12 for mRNA delivery with lipidoid OF-02 (cKK core with 4 non-cleavable 1,2-amino alcohol linoleyl tails), which has a near identical liver-only biodistribution but significantly higher in vivo mRNA translation of hEPO compared to cKK-E12 [153]. Replacing the OF-02’s non-cleavable tails with ester led to OF-Deg-Lin, which dramatically changed the in vivo mRNA delivery to specifically targeting the spleen [154]. Other functional lipidoid scaffolds include the phenyl core FTT5 with hyper-branched tails [155], dendrimer-like 5A2-SC8 [156], polymer-based 7C1 [157], aminoglycosides-based GT-EP10 [158], etc.
4. Organ-specific and cell-specific RNA delivery via targeted LNPs
4.1. Early development of organ-specific and cell-specific liposomal delivery systems
As the early generation of “organ-specific” delivery systems, liposomes are typically biodistributed to the liver and spleen, i.e., the primary organs associated with the reticuloendothelial system (RES)/mononuclear phagocyte system (MPS). The PEGylation of liposomes delayed the uptake by the RES/MPS and prolonged blood circulation [90]. Ultimately, PEGylated liposomes shared biodistribution similar to that of non-PEGylated liposomes, which accumulate in the liver and spleen after intravenous injection [52,159]. Unfortunately, compared to the EPR-driven liposomal PK, antibody-liposome conjugates failed to demonstrate additional PK benefits for tumor targeting [160]. Increasing the particle size of liposomes is known to increase spleen uptake [161]. It is well-known in the 80s that liposomal surface charges are known to impact its pharmacokinetics [162]; for example, increasing cationic lipids, e.g., DOTAP and DOTMA, in the liposomal formulations strongly enhance lung accumulation [163–165].
Further, sending liposomes to targeted cells has always been the goal at the birth of liposomal drug delivery in the 70s. Gregoriadis and Neerunjun explored the use of non-covalently functionalizing liposomes with IgG as a strategy for homing bleomycin-encapsulating liposomes to target specific cells in 1975 [166]. Magee et al. used IgG-functionalized liposomes for targeted delivery of “immune RNA” to lymphocytes for immune stimulation in 1978 [167]. Covalently functionalized antibody-liposome conjugates were explored for target delivery to myocardial infarction sites by Torchilin et al. in 1979 and for cell-type specific targeting by Huang et al. in 1980 [168,169]. Antibody-liposome conjugates were the major way to achieve site-specific/organ-specific therapeutic delivery, as demonstrated by Holmberg and Leaf Huang et al. in 1990 [170]. Reported in the same year by Klibanov and Leaf Huang et al., PEGylated liposomes significantly prolonged blood circulation [90], facilitating the tumor Enhanced Permeation and Retention (EPR) effect, which eventually took over the major focus for developing liposomal formulations for cancer delivery following the approval of Doxil® in 1995 [91]. On the other hand, antibody-drug conjugates are still being developed as the major targeted drug delivery platform for cancer to this date. Compared to liposomal delivery systems, antibody-drug conjugates are limited to very low drug loading capacity and cannot provide barrier protection for therapeutic RNAs from enzymatic degradation [171]. Thus, there is still a need to develop site-specific LNP systems for precision RNA therapeutics.
4.2. Liver/spleen-, liver-, spleen-, and lung-specific LNPs
At the front end of this development are the selective organ targeting (SORT) LNPs developed by Cheng and Siegwart et al. using ionizable lipids 5A2-SC8, MC2, and lipidoids C12–200. By adjusting the ratio of supporting lipids such as DOTAP or 18PA, the SORT LNPs could achieve exclusive organ-targeted mRNA delivery to the lung (with 50% DOTAP), liver (<10% DOTAP), and spleen (10–15% DOTAP with 10–40% 18PA). The ability of SORT LNPs to achieve organ-specific mRNA delivery is proposed for targeted CRISPR-Cas gene editing [172,173], which has led the phase 1 clinical trials of CT1100 (Lung SORT LNPs by ReCode Therapeutics) for Cystic Fibrosis in February 2024. LoPresti et al. reported a variation of the SORT LNPs using the ionizable lipid 304O13. Liver-favored distribution was observed when co-forming LNPs with the neutral lipids 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), sphingomyelin (SM), and ceramide (Cer). Spleen-favored distribution was achieved when co-formulating LNP formulations with zwitterionic lipid phosphatidylserine (PS) and anionic lipids phosphatidylglycerol (PG) and phosphatidic acid (PA). Cationic DOTAP and ethyl phosphatidylcholine (EPC) shifted the LNP distribution to the lungs [174]. When delivered via intratracheal (IT) administration, MC3 and DOTAP co-formulated LNPs have been shown to effectively deliver mRNA to alveolar epithelial cells (AECs) and fibroblasts in fibrotic lungs [175].
Similar to liposomes, LNPs are PEGylated and typically exhibit liver/spleen distribution following systematic administration. Leveraging liver biodistribution, Onpattro® LNPs (ionizable lipid = MC3 with non-cleavable lipid tails) are designed to treat liver hATTR amyloidosis [123,176]. The high liver accumulation of MC3 LNPs sparks the development of ionizable lipids with cleavable ester tails. Ester lipid tails have been used in clinically approved SM-102, ALC-0315 ionizable lipids, and many other structural derivatives sharing liver biodistributions for improving the biodegradability of ionizable lipids in liver [177]. On the lipidoid side, the classic lipidoid 5-tail piperazine-based C12–200 is known to target both liver and spleen when given intravenously [177]. The 4-tail piperazine-based 246C10, however, showed liver-only distribution in a CRISPR/Cas gene editing study targeting antithrombin (mAT) for treating hemophilia [11]. With 2 extra carbons in the lipid chain in 246C10, LNPs based on piperazine-based IC8 (246C12) were shown to distribute to the liver and spleen when delivering mRNA encoding B7H3 × CD3 bispecific T-cell engagers (BiTEs) antibodies. The liver was targeted as an in vivo antibody factory for BiTEs expression and secretion to treat hematologic malignancies and melanoma [178]. LNPs based on modified diketopiperazine (DKP)-based lipidoids, e.g., cKK-E12 (non-cleavable saturated tails) and OF-02 (non-cleavable unsaturated tails), showed a highly liver-focused distribution with higher protein translation efficiency by OF-02 compared to cKK-E12 [153]. Interestingly enough, by modifying DKP-lipidoids with cleavable ester and unsaturated tails, e.g., OF-Deg-Lin, although still distributed to the liver more than the spleen, the spleen accounted for over 85% of the mRNA expression [154]. Incorporating oleic acid (OA) in LNP formulations significantly enhanced liver distribution [164], whereas replacing cholesterol with cholesteric acid enables LNPs to exhibit a splenic tropism [112]. Additionally, incorporating different helper lipids in LNPs is known to alter their interaction with serum apolipoprotein E (ApoE). Having DOPE in the LNP formulation leads to higher ApoE binding and enhances liver delivery. Replacing DOPE with DSPC decreases ApoE binding, reducing liver uptake and increasing spleen distribution [179].
4.3. Placenta and fetus distribution of liver/spleen targeted LNP during pregnancy
Neither the liver-targeting LNPs formulated by DKP-lipidoids, e.g., cKK-E12 nor OF-02, nor the classic liver-and-spleen targeting lipidoids C12–200 distributed to the uterus/ovaries after systematic distribution [154]. By modifying the lipid tails of C12–200 with ether linkages, ionizable lipids A-4 showed an over 20× distribution to the spleen over the liver in non-pregnant mice. More interestingly, when given systematically in pregnant mice, LNPs A-4 are highly efficient in biodistribution to the placenta but not the fetus. A similar effect was observed on a similar ionizable lipidoid derivative B-5, which also has an ether structure [177]. This phenomenon, where highly spleen-targeted LNPs breaching into the placenta during pregnancy, is proposed for delivering mRNA-VEGFR to increase placenta blood flow for placental insufficiency. Whether this pregnancy-dependent biodistribution applies to other spleen-targeted LNPs, such as OF-Deg-Lin, remains to be investigated, and this unique observation/demonstration also highlighted the importance of developing precision RNA delivery to minimize undesirable off-target effects. Finally, in-utero delivery of LNP A-4 (PA4) was able to deliver mRNA more efficiently to the liver, lung, and intestine of the fetus with minimal fetal immunotoxicity or liver damage compared to mRNA delivered by MC3-LNP or jetPEI cationic polymer [146]. Meanwhile, the MC3 LNPs were shown to deliver mRNA to the heart, diaphragm, and skeletal muscle, in addition to the liver of the fetus via in-utero delivery [180].
4.4. Pancreatic distribution of liver-and-spleen targeted LNP via intraperitoneal injection
The administrative route is known to significantly impact the pharmacokinetic and protein translation half-live of mRNA-delivering LNPs [181]. When given intravenously, liver-targeting lipidoids 306Oi10 (ester lipid tails, 98.6% liver), 200Oi10 (a C12-C200 derivative with ester tails, 97.7% liver), and liver-and-spleen-targeting 514O6,10 (branched ester lipid tail, 68.3% liver and 24.6% liver) showed typical LNP biodistribution. However, given via intraperitoneal injections, significantly increased pancreas distribution was observed for 306Oi10 (14.7%), 200Oi10 (46.4%), and 514O6,10 (52.3%). This pancreatic distribution could be further enhanced by incorporating cationic lipid DOTAP in the LNP formulation, which universally increases pancreas mRNA expression for all three lipidoids tested [182].
4.5. LNPs for bone microenvironment RNA delivery
Bisphosphonate (BP) ligands, such as alendronate and pamidronate, have long been studied as drug conjugates for targeting metabolic bone diseases [183]. BP-lipid conjugates have been used for bone-targeting liposomal delivery of doxorubicin [184]. Similarly, BP-conjugated lipidoids 490BP-C14, while majorly accumulating in the liver and spleen after IV injections, were able to distribute to the bone microenvironment in mice and demonstrate mRNA-EGFP expression in the femur [185]. For genome editing therapy targeting haematopoietic stem cells (HSCs), incorporating covalent crosslinker/lipid crosslinkers (e.g., AA11, a 3-sulfo-N-hydroxysuccinimide modified fatty ester) in the LNP formulations has also been demonstrated to alter the serum protein coating on the LNP (also known as the protein corona), which led to bone marrow homing at 20% molar ratio in 5A2-SC8-based LNPs [186].
4.6. Eye-specific and ear-specific RNA delivery via localized LNP administrations
Represented by the FDA-approved adeno-associated virus (AAV)-based gene therapy, Luxturna™, eye-specific gene delivery via subretinal delivery has been widely explored for treating inherited retinal degeneration/blindness and other retinal degeneration diseases [187]. Using MC3/KC2 formulated LNPs, mRNA given via subretinal delivery was able to demonstrate gene expression in the eyes for 5 days, paving new ways for repeatable non-immunogenic gene therapy in the eyes [188].
On the other hand, while no FDA-approved gene therapies exist, AAV-gene therapy is being studied in clinical trials to restore hearing in autosomal recessive deafness [189,190]. Cationic lipid-mediated in vivo delivery of Cas9–guide RNA complexes via microinjection to the inner ear has shown great promise as a non-viral genome editing therapy in preclinical studies [191].
4.7. Immune cell-targeting LNPs for immunoengineering
Immunoengineering is a dynamic and growing field that leverages the immune system’s capabilities to combat diseases, notably cancer and autoimmune disorders. LNPs have revolutionized this field by offering precise delivery of immunomodulatory molecules, transforming immune cells into precise, targeted therapeutic agents. An essential area of focus is the creation of tolerogenic vaccines that promote immune tolerance, potentially transforming treatments for autoimmune diseases such as multiple sclerosis [16], preventing anaphylaxis for peanut allergies [15], and allergic airway disease [192,193]. These vaccines function by inducing antigen-specific regulatory T cells (iTregs). LNPs serve as delivery vessels, carrying mRNA that encodes the target antigens or allergens to specific antigen-presenting cells, such as liver sinusoidal endothelial cells (LSECs) or tolerogenic dendritic cells [194]. By employing mannose ligands that bind to the mannose receptor CD206 on LSECs and optimizing the LNP composition with PEGylated lipids, this mannose-mediated approach efficiently homes in on the target cells. This strategy, utilizing liver-targeting ionizable lipids like 246C10 and MC3 for LSEC targeting, has been validated in several studies [15,145,195]. Inflammatory diseases have also been addressed using LNPs. For instance, Veiga et al. attached anti-Ly6c antibodies to LNPs to home in on and treat Ly6c + inflammatory leukocytes involved in inflammatory bowel disease [196]. In cardiology, Rurik et al. utilized LNPs outfitted with anti-CD5 antibodies to target T cells in the spleen. These modified T cells are then reprogrammed into CAR-T cells that target the fibroblast activation protein (FAP), offering a novel strategy to treat cardiac injury [197].
5. Barcoding technologies for high-throughput LNP formulation discovery
As mentioned earlier, gene editing by CRISPR-Cas-encoding mRNA is a burgeoning application of RNA therapeutics, driving the ever-increasing demand for cell-specific precision RNA delivery. This also means a significantly increased need to work with genetically modified transgenic animal models when developing such LNP formulations. Barcoded LNP formulations are becoming essential for developing next-generation gene editing therapeutics to facilitate LNP discovery with reasonable efficiency and feasibility [198].
5.1. DNA-encoded library (DEL) for high-throughput screening (HTS)
DNA-encoded library (DEL) is commonly used in fragment-based drug discovery, e.g., to discover proteolysis targeting chimeras (PROTACs) by screening for the protein binding of billions of compounds in one microcentrifuge tube. The DEL-enabled high-throughput screening (HTS) could be done in a few days instead of years when using the traditional drug screening method in microtiter plates. This is achieved by attaching every drug fragment (experiment variables) to a unique DNA sequence, i.e., the barcode [199]. The biggest advantage of the DEL/DNA-barcoding approach is that it allows sample pooling. In the context of PROTACs discovery, 1E4-1E12 DNA-tagged molecules could be pooled in one microtube for protein binding studies [200]. Non-binding molecules are washed away along with their DNA barcodes, and the remaining DNA barcodes of the “hit” molecules that bound to the target protein can then be amplified and tagged for DNA sequencing using standard polymer chain reactions (PCR) workflow. The identity of the drug candidates could then be “decoded” by next-generation DNA sequencing and ranked by their abundance to identify the top candidates/DNA barcode. Compared to traditional HTS drug screening, the DEL/DNA-barcoding approach could significantly reduce the time from years to days and resources required, allowing HTS screening in vitro and in vivo via sequencing.
5.2. Tracking LNPs in vivo with molecular tracers and DNA barcodes
In drug delivery system development, molecular tracers are instrumental in conducting pharmacokinetic (PK) and pharmacodynamic (PD) studies. As shown in Fig. 5a–d, these tracers allow for the tracking of LNPs at resolutions that range from whole-body to single-cell levels, each requiring specific probes for accurate localization [201]. Radioisotopes provide the best tissue penetration length and exceptional signal-to-noise ratios, which are directly translated from preclinical animal models to human studies. Through 14C and 3H labeling, LNPs can be used to radiolabeled without modifying chemical structures [101,120,202,203]. Alternatively, chelator-conjugated lipids, e.g., DSPE-DTPA, could also be incorporated into LNP formulations for PK studies. However, radiation-based tracers do not provide sufficient spatial resolution down to tissue and cellular levels, which are centric to gene delivery studies. Fluorescent imaging with lipid-conjugated dyes—such as DiI, DiO, DiD, DiA, and DiR—is advantageous for higher spatial resolution at the organ, tissue, and cellular levels. These dyes allow researchers to investigate the spatial distribution and cellular uptake of LNPs in small animal models, although their shallow tissue penetration limits their use [52,204]. However, tracking LNPs alone does not confirm the successful delivery and function of their cargo—mRNA or siRNA—within the target cells. To assess this directly, fluorescent reporter proteins like GFP, RFP, and mCherry are common tools for evaluating protein synthesis following mRNA delivery or for monitoring siRNA silencing efficiency by tracking the loss of fluorescence in genetically modified reporter cells. For a more dynamic and amplified signal, mRNA encodes bioluminescent proteins, such as firefly luciferase (fLuc) is commonly used. The enzyme fLuc catalyzes the emission of light in the presence of its chemical substrate, luciferin, enabling a single mRNA translation event to produce a substantially heightened luminescent output, offering a stark contrast to the one-to-one signal ratio seen with fluorescent proteins from mRNA translation. However, the bioluminescent method is restricted to living cells, as the enzymatic reaction requires a live cell environment (Fig. 5c). To bypass this limitation and to boost the visibility of successful mRNA delivery, especially at low delivery efficiencies, transgenic animals are engineered with a genetic control system for reporter protein expression. For example, transgenic mouse models such as Ai9LSL-tdTomato and Ai14LSL-tdTomato carry a tdTomato (a red fluorescent protein) reporter gene preceded by a loxP-flanked STOP cassette, i.e., the reporter gene is initially silenced. The administration of LNPs containing Cre recombinase mRNA to these mice triggers the expression of Cre in target cells. Cre recombinase then excises the STOP cassette, thus activating reporter tdTomato gene expression. The resulting bright red fluorescence serves as a robust and precise marker of successful Cre mRNA delivery and expression. This sophisticated strategy allows for the signal amplification necessary for analysis, even in fixed tissues or cells, as illustrated in Fig. 5d [205].
Fig. 5. Biodistribution tracers for developing organ-specific and cell-specific LNPs.

a. Spatial resolution of commonly used molecular tracers for PK/PD studies. b. Molecular labeling techniques for tracking lipids in the LNPs. c. Comparing bioluminescent-based and fluorescent-based imaging techniques for tracking RNA delivery by LNPs. d. Illustration of using Cre-Lox system for detecting mRNA delivery. e. The DNA-barcoding workflow for HTS LNP formulation screening in vivo. (panels and d are partially created with BioRender.com).
When DNAs are used as the molecular tracer, DNA “barcodes” could be formulated with the ionizable LNPs, where each varying formulation composition is designated with a unique DNA sequence (unnatural sequence) that is sandwiched with PCR primer binding sites (Fig. 5e). Similar to the DEL HTS screening, hundreds to thousands of DNA-barcoded LNPs could then be pooled into one single injection for evaluating the pharmacokinetics of LNP in one single animal [206]. After the experiment, the organ-specific or cell-specific biodistribution of LNP-delivered DNA barcodes could be extracted and amplified by PCR. By fusing PCR primers with next-generation sequencing (NGS)-enabling tags, the amplified DNA barcodes could then be sequenced and ranked by abundance at tissue-, organ-, and cellular levels, representing the biodistribution ranking of the barcoded LNP formulations. Using DNA barcoding with just 8 nucleotides, 4E8 combinations could be generated, equating to 65,536 sequence-unique DNA barcodes. Massive LNP formulations could be screened with minimal animal numbers, thus enabling HTS LNP formulation discovery in transgenic models that are not practically feasible using other molecular tracers.
5.3. Standard-, enhanced-DNA-barcoding and mRNA/peptide-barcoding strategies for LNPs
Dahlman et al. demonstrated the use of DNA-barcoded LNPs in 2017 by screening 30 siRNA-delivering LNP formulations per one mouse to identify the optimal PEG-lipid tail length, PEG molecular weights, and PEG % for delivery to the brain, heart, kidney, liver, lung, skeletal muscle, uterus, and pancreas, using LNPs based on lipidoids 7C1 and C12–200 [207]. Huayamares et al. screened 94 DNA-barcoded mRNA-LNP (7C1) formulations in one injection per animal to develop liver “de-targeted” formulation for enhancing mRNA delivery to a solid tumor model of neck squamous cell carcinoma (FaDu xenograft/HNSCC) [208]. While DNA-barcoding is extremely powerful for in vivo HTS of LNP formulations, detecting DNA barcodes reflects only the distribution of the LNP, i.e., the pharmacokinetics of the LNPs. For RNA therapeutics, delivering to the targeted organ or cells, however, does not equate to mRNA translation or siRNA gene silencing due to the endosomal escape barriers mentioned above. In other words, using DNA barcoding itself is not enough to evaluate the efficacy and pharmacodynamics of RNA therapeutics. Therefore, co-delivery of reporter RNA molecules is often advised to establish the PK/PD profile of RNA-delivering LNP formulations. For example, Sanchez et al. utilized 128 LNP formulations co-encapsulating DNA-barcode/Cre-mRNA using stereo-pure R- or S-C12–200 and racemic C12–200. By pooling 55 formulations/injection per Ai9/Ai14LSL-tdTomato mouse, successful mRNA delivery and translation can be identified by isolating tdTomato-positive cells following NGS sequencing to decode the LNP formulation. The S-configured C12–200 was found to significantly enhance mRNA translation compared to the R-configured C12–200 [209]. Similar strategies have been demonstrated by Lokugamage et al. screening 82 cKK-E12-based LNP formulations co-encapsulating DNA-barcodes and Cre-mRNA, per Ai14LSL-tdTomato mouse, for identifying LNPs targeting liver Kupffer cells, endothelial cells, and hepatocyte [210]. Ni et al. investigated 128 structurally different ionizable lipidoids sharing a piperazine core and using Cre-mRNA/DNA barcoding to pool-inject 65 LNP candidates per Ai14LSL-tdTomato mouse, which led to the discovery of PPZ-A10 (Pi-A10) as a novel mRNA-delivering lipidoid targeting hepatic and splenic immune cells (Fig. 4g) [149].
In addition to NGS-based barcoding strategies, HTS proteomic profiling via LC-MS/MS provides an alternative avenue for evaluating mRNA-delivering LNPs in vivo. As demonstrated by Rhym et al., this is achieved by fusing reporter mRNA with a peptide-barcoding sequence, that both the reporter gene and the peptide-barcodes are detectable when mRNA delivery and translation are both successful [211]. Using mRNA barcoding, Rhym et al. evaluated 384 structurally different ionizable lipids in only 9 mice, identifying RM133–3 (Fig. 4f) as a novel ionizable lipid for mRNA delivery [127]. All considered, the combined use of fluorescent/bioluminescent molecule probes, standard-, enhanced-DNA-barcoding, and mRNA/peptide-barcoding strategies provide HTS LNP screening with precision and resolution down to single cell levels, paving new ways of fast-tracking organ-specific and cell-specific LNP for prevision RNA delivery, as summarized in Table 1.
Table 1.
LNPs for organ-specific and cell-specific RNA delivery.
| Targeted Organ(s) or Cell(s) | Barcoding (Y/N) | Ionizable Lipids (% molar ratio) | Supporting Lipids & Targeting Ligands | RNA Cargo | Animal | Proposed Applications | Ref. |
|---|---|---|---|---|---|---|---|
| Liver | MC3 50% |
DSPC, lipid-PEG and β-sitosterol | mRNA | BALB/c mice | SARS-CoV-2 | [257] | |
| Liver | MC3 | DOPE, lipid-PEG and Chol | mRNA | C57BL/6 J mice | Liver cancer | [258] | |
| Liver | OF-02 35% |
DOPE, lipid-PEG and Chol | mRNA | C57BL/6 mice | Protein/enzyme replacement therapy | [153] | |
| Liver | G0-C14 5% |
PDSA and lipid-PEG | mRNA | Nude, BALB/c, C57BL/6 mice | Liver cancer | [259] | |
| Liver & Spleen | IC8 35% |
DSPC, lipid-PEG and Chol | mRNA | NSG mice | hematologic malignancies and melanoma | [178] | |
| Liver | 246C10 26.5% |
DOPE, lipid-PEG and Chol | mRNA sgRNA |
C57BL/6 mice | Gene editing Hemophilia |
[11] | |
| Liver | 5A2-SC8 24% |
DOPE, lipid-PEG and Chol | mRNA sgRNA siRNA |
C57BL/6 mice | Gene editing Liver cancer |
[156] | |
| Liver | GT-EP10 (A15) 20% |
DOPE, Chol, DMG-PEG2kDa | mRNA | C57BL/6 mice | Protein/enzyme replacement therapy | [158] | |
| Liver | 5A2-SC8 39.5% |
DODAPa, DOPE, lipid-PEG and Chol | |||||
| Lung | 5A2-SC8 11.9% |
DOTAPb, DOPE, lipid-PEG and Chol | mRNA sgRNA |
C57BL/6 mice, Ai14LSL-tdTomato mice |
Gene editing | [173] | |
| Spleen | 5A2-SC8 16.67% |
18PAc, DOPE, lipid-PEG and Chol | |||||
| Liver | DOPE, C14-PEG and Chol | ||||||
| Lung | 304O13 35% |
DOTAP, C14-PEG and Chol | mRNA | C57BL/6NCrl mice | Multiple | [174] | |
| Spleen | PC, C14-PEG and Chol | ||||||
| Muscle | TCL053 60% |
DPPC, lipid-PEG and Chol | mRNA sgRNA |
hEx45KI-mdx44 mice | CRISPR-Cas gene editing (Duchenne muscular dystrophy) | [251] | |
| Bone | No | 490BP-C14 35% |
DOPE, lipid-PEG and Chol | mRNA | C57BL/6 J mice | Skeletal diseases | [185] |
| Bone | 5A2-SC8 19% |
DOPE, Chol, DMG-PEG2kDa, Crosslinker (20%) |
mRNA | C57BL/6 & Ai14LSL-tdTomato mice | HSCs gene editing | [186] | |
| Pancreases (via IP injection) | 306Oi10 200Oi10 (2nd best) 514Oi6, 10 (the best) 35% |
Chol, C14-PEG2kDa Helper: DOTAP (better) or DOPE or PS |
mRNA | C57BL/6 mice | Diabetic, Pancreatic cancer |
[182] | |
| Lung alveolar epithelial cells (and fibroblasts (intratracheal) | MC3 10–25% |
DOTAP (40%) DPPC, Chol, DSPE-PEG2kDa |
mRNA | C57BL/6 mice | Idiopathic pulmonary fibrosis | [175] | |
| Lung (via inhalation) | IR-117–17 (A10-Lin) IR-19-Py 50.1% |
DOTAP, Chol, C14-PEG2kDa | mRNA | C57BL/6 J B6.Cg-Gt(ROSA) 26Sortm14(CAG-tdTomato)Hze/J C57BL/6 N Scnn1b-Tg |
Lung diseases (e.g., cystic fibrosis, primary ciliary dyskinesia, α-1 antitrypsin deficiency, vaccination, and asthma | [22] | |
| Fetal Liver (in utero) | PA3-C12/C14 (A-3/B-3) 35% |
DOPE, Chol, C14-PEG2kDa | mRNA | Balb/c | Protein/enzyme replacement therapy | [146] | |
| Placenta (IV injection) | A-4 35% |
DOPE, Chol, C14-PEG2kDa | mRNA (VGEF-A) |
Pregnant mice: gestational age E16 | Placental insufficiency | [177] | |
| Spleen | mRNA | Non-pregnant mice | |||||
| Retinal | MC3, KC2 50% |
DSPC, DMG-PEG2kDa DSPC, PEG-lipid and |
mRNA | Balb/c | Retinal degeneration | [188] | |
| Leukocyte | MC3 50% |
Chol Anti-Ly6c Abs |
mRNA | C57BL/6 mice | Inflammatory Bowel Disease | [196] | |
| T cells (Spleen) | L319 55% |
DSPC, PEG-lipid and Chol Anti-CD5 Abs, |
mRNA | C57BL/6 N, Ai6LSL-ZsGreen1 mice | Cardiac Fibrosis | [197] | |
| LSECs | MC3 50% |
Mannose, DSPC, PEG-lipid and Chol + Mannose | mRNA | C3H/HeJ mice | Tolerogenic vaccine Peanut Allergy | [15] | |
| LSECs | 246C10 26.5% |
DOPE, PEG-lipid and Chol + Mannose | mRNA | C57BL/6, Ai14LSL-tdTomato mice | Tolerogenic vaccines | [145] | |
| SECs/LSECs | MC3 50% |
DSPC, PEG-lipid and Chol | mRNA | C57BL/6 mice | Tolerogenic vaccines | [195] | |
| B cells | OF-Deg-Lin 35% |
DOPE, PEG-lipid and Chol | mRNA | C57BL/6 mice | ImmunoEngineering | [154] | |
| Liver | C12–200 50% |
DSPC, lipid-PEG and Chol | siRNA | C57BL/6 mice | Multiple | [207] | |
| Lung | 7C1 70% Stereo-pure |
lipid-PEG and Chol | |||||
| Liver | C12–200- based 40% |
DOPE, lipid-PEG and 20α-OH | mRNA | Ai14LSL-tdTomato mice | Protein/enzyme replacement therapy | [209] | |
| Tumor | 7C1 50% |
DSPC, lipid-PEG and Chol | mRNA | FaDu xenograft nude mice | Head and neck cancers | [208] | |
| Lung | 3-A2–7b 35% |
DOPE, Chol, C14-PEG2kDa | mRNA-Cas9 sgRNA-VEGFR2 |
Ai14LSL-tdTomato, C57BL/6 J mice | CRISPR-Cas gene editing (e.g., lung cancer) | [20] | |
| Lung ECs | 7C1 50% |
18:1 Lyso PC, PEG-lipid and Chol | mRNA-SpCas9, sgRNA, siRNA | C57BL/6 J mice | CRISPR-Cas Gene editing for cystic fibrosis |
[260] | |
| Spleen ECs | 7C1 60% |
DOPE, PEG-lipid and Chol | mRNA | Ai14LSL-tdTomato mice | ImmunoEngineering | ||
| Myeloid progenitors | Yes (DNA) |
7C1 60% |
PEG-lipid and Chol | N/A | C57BL/6 J mice | NA | [261] |
| Liver ECs | 7C1 50% |
18:1 Lyso PC, lipid-PEG and cholesteryl oleate | mRNA-SpCas9, sgRNA |
C57BL/6 J mice (WT, LDLR−/−, VLDLR−/−) | CRISPR-Cas gene editing | [262] | |
| BMECs, Lung and spleen | 7C1-based 80% |
PEG-lipid | siRNA and Bortezomib | C57BL/6 mice, MM.1S-xenograft NOD SCID mice | Multiple Myeloma | [263] | |
| Liver ECs and KCs; Lung ECs | Stereo-pure 7C1-based 80% |
PEG-lipid and Chol | siRNA | C57BL/6 J mice (WT, ApoE−/−, LDLR−/−, and VLDLR−/−) | NA | [264] | |
| KCs | cKK-E15 45% |
DOPE, PEG-lipid and Chol | mRNA | Ai14LSL-tdTomato mice | ImmunoEngineering Vaccination |
[210] | |
| Lung ECs | cKK-E12 35% |
18:0 DDAB, PEG-lipid and Chol | mRNA, | Ai14LSL-tdTomato mice, C57BL/6 J mice |
Gene editing. | [265] | |
| Liver KCs and ECs | cKK-E12 50% |
DOPE, PEG-lipid and 20α-OH | mRNA | Ai14LSL-tdTomato mice | ImmunoEngineering Vaccination |
[266] | |
| Liver KCs Spleen macrophages | PPZ-A10 35% |
DOPE, Chol, C18-PEG2kDa | mRNA | Ai14LSL-tdTomato mice | ImmunoEngineering Vaccination |
[149] | |
| Lung ECs | RCB-4–8 30% |
DOTAP, Chol, C14-PEG2kDa | mRNA-SpCas9 sgRNA |
C57BL/6, Ai9LSL-tdTomato mice | CRISPR-Cas gene editing for cystic fibrosis | [21] | |
| Liver, spleen, lung | Yes (mRNA) |
C12-200 35% (liver, spleen) 60% (lung) |
DOPE, Chol, C14-PEG2kDa | mRNA | C57BL/6 | CRISPR-Cas9 gene editing, vaccines, immunoRx | [267] |
| Liver | RM-133-3 35% |
DOPE, lipid-PEG and Chol | mRNA | C57BL/6 mice | Protein Rx (e.g., hEPO) | [127] |
6. Milestones and clinical landscape in the development of mRNA and siRNA therapeutics
6.1. A six-decade-long journey from discovery to clinical approvals of mRNA and siRNA therapeutics
Parallel to the discovery of liposomes, Sydney Brenner, François Jacob, and Matt Meselson discovered the role of mRNA in 1961 [212–214]. As an intermediate between DNA and protein translation, the discovery of mRNA and ways to synthesize stabilized mRNA (inclusion of PolyA tails and RNA capping technologies for enhanced stabilities and translation efficiency) for in vitro/in vivo protein expression alongside the gene delivery endeavors via liposomal formulations collectively embarked on a six-decade-long journey in developing mRNA therapeutics (Fig. 6 and Table S2) [215–219]. Therapeutic applications of mRNA were explored in various directions in the ‘90s when Wolff et al. first demonstrated in 1990 using mRNA for direct in vivo gene transfer and protein expression in mouse skeletal muscle [219]. The successful use of mRNA for in vivo protein translation led to the exploration of mRNA-mediated diabetes treatments by Jirikowski in 1992 [220], anti-influenza cytotoxic T-cell induction using mRNA-liposomes by Martinon et al. in 1993 [221], mRNA cancer vaccines by Conry et al. in 1995 [222], mRNA-enabled dendritic cell vaccine by Boczkowski et al. in 1996 [223], mRNA-mediated overexpression of receptor in mammalian cells (conceptionally leading to modern-day CAR-T engineering via mRNA-LNP) by Karikó et al. in 1999 [224], and mRNA melanoma vaccine for cytotoxic T cell induction by Zhou et al. in 1999 [225] among many other mRNA-enabled vaccination studies [222,223]. More importantly, Katalin Karikó and Drew Weissman discovered that modified nucleoside significantly reduced the innate immune response to exogenous mRNA, enhancing its stability and efficacy [226]. Such advancements proved instrumental fifteen years later when mRNA technology was rapidly applied to develop mRNA-based COVID-19 vaccines. The discovery was later awarded the Nobel Prize in Physiology in 2023 [227]. On the other hand, siRNA was discovered for posttranscriptional gene silencing in 1999 by Hamilton and Baulcombe et al., nearly four decades after mRNA was discovered [228]. As shown previously in Fig. 1, siRNA functions as a targeted mRNA degrader, leading to targeted gene silencing. The unique mechanism of siRNA led to their first approval for clinical use in 2018 as Patisiran, an MC-3 lipid-formulated siRNA that silences the amount of wild-type and mutant transthyretin mRNA for treating hereditary transthyretin-mediated amyloidosis [29,229]. This culmination of decades of RNA research and innovation in RNA-LNP delivery systems collectively set the stage for the diverse clinical landscape of mRNA and siRNA therapeutics we see today.
Fig. 6. A six-decade-long journey from discovery to clinical approvals of mRNA and siRNA therapeutics.
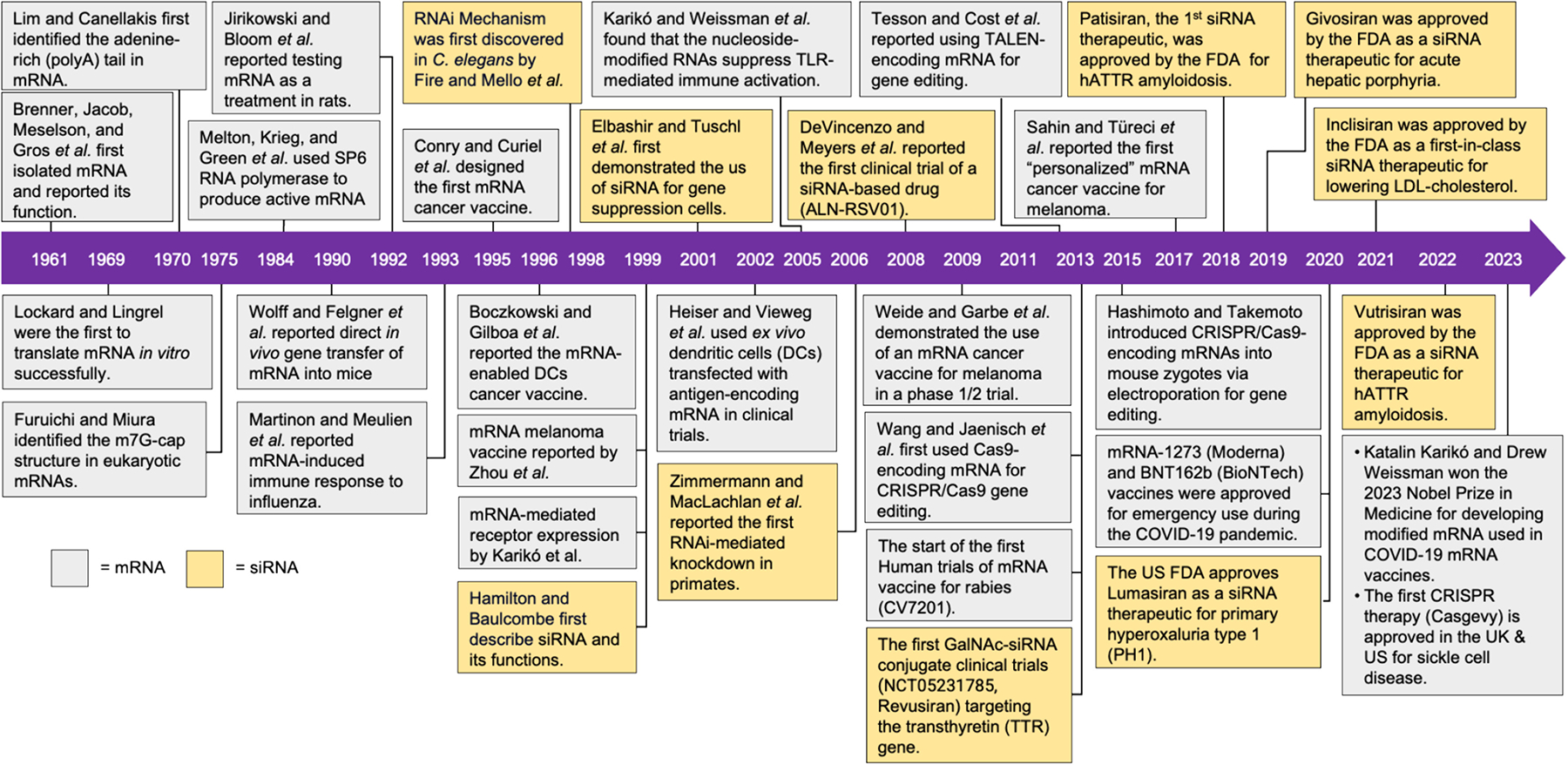
This retrospective timeline summarizes the development of mRNA and siRNA therapeutics, with selected milestones highlighting key developments for enabling their clinical applications. Full itemized information is summarized in Table S2.
6.2. The clinical landscape of mRNA and siRNA therapeutics
The burgeoning field of mRNA and siRNA therapeutics has transitioned from foundational research to clinical applications with unprecedented speed. The clinical landscape today is diverse, encompassing vaccines, gene editing, and gene silencing approaches that tackle a broad range of diseases. In the vanguard of mRNA therapeutics are the COVID-19 vaccines, Tozinameran and Elasomeran, known commercially as BNT162b2 and mRNA-1273. These vaccines utilize LNPs as delivery systems, successfully stimulating protective immune responses against SARS-CoV-2. Their development highlights the agility of mRNA platforms in adapting to pathogenic mutations, a contrast to traditional vaccine development methods. This flexibility was demonstrated when Moderna’s mRNA-1273 entered clinical trials within two months of the viral genome’s publication [230].
Beyond its pivotal role in combating COVID-19, mRNA therapeutics are expanding their reach to target a range of formidable infectious diseases [231]. Clinical trials are currently underway for next-generation vaccines aimed at preventing influenza, HIV/AIDS, and rabies [232–236]. Meanwhile, CRISPR-Cas9 gene editing has also advanced through the use of mRNA delivery. Targeted gene editing is achieved by encapsulating Cas9 mRNA and sgRNA within LNPs [9]. This innovative approach is now being tested in clinical settings. A phase I trial for gene editing in patients with Primary Ciliary Dyskinesia (RCT1100, NCT05737485, targeting the DNAI1 gene) and another for Cystic Fibrosis (RCT2100, NCT06237335, targeting the CFTR gene) are noteworthy. Both trials have dosed the first patient in 2024 via inhaled LNPs for lung delivery.
The realm of siRNA therapeutics is equally diverse and has been applied to a variety of cancers, including lung cancer, pancreatic cancer, breast cancer, prostate cancer, ovarian cancer, and liver cancer, among others [237–242]. Tailored siRNA molecules enable the targeting of specific cancer gene sequences, offering personalized treatment options. The versatility of siRNA is not limited to cancer; it has emerged as a powerful tool against genetic and metabolic diseases (Table S3) [29,49].
An overview of the current clinical trials in mRNA and siRNA therapies is distilled from an extensive search of ClinicalTrials.gov (Fig. 7a). These trials are categorized into mRNA and siRNA therapeutics, with the mRNA trials further clustered as (i) mRNA therapeutics (which imparts therapeutic action) and (ii) mRNA-enabled cellular therapy (where mRNA-encoded proteins/receptors/ligands augments cell-based treatments) [243]. As shown in Fig 7, b1, mRNA therapeutics have reached clinical approval through the mRNA COVID-19 vaccines, with highly active pipelines across phase I-III. Diving into the indications of mRNA therapeutics, the majority of the trials are COVID-19 related, as expected, followed by the up-and-rising applications such as mRNA-vaccine for infectious diseases, oncology, genetic disorders, cardiovascular, etc. (Fig. 7, b2). Diving into the oncology-related indications, the top application is evidently the personalized mRNA cancer vaccines, followed by other indications in gastrointestinal, melanoma, liver cancer, pancreatic cancer, and others (Fig. 7, b3). As for infectious diseases, the leading applications are influenza vaccines (~45%), followed by Respiratory Syncytial Virus (RSV) vaccines, HIV vaccines, cytomegalo-virus (CMV) vaccines, Rabies vaccines, and others (Fig 7, b4).
Fig. 7. Clinical Landscape of mRNA and siRNA Therapeutics.

A. Search terms for clinicaltrial.gov using keywords included “siRNA,” “mRNA,” “RNAi,” “messenger RNA,” and “interfering RNA.”. Exclusions were made for trials that did not involve mRNA or siRNA as therapeutic— remaining trials were included for further analysis (detailed in Fig. S1). B. mRNA therapies, C. mRNA/cell therapies, and D. siRNA therapies. E. Analyzing indications in mRNA therapy, the top 3 indications were classified by phase and marked in F. Conducting a similar analysis for mRNA/cell therapy and siRNA therapy, with mRNA/cell therapy labeled in G. H and siRNA labeled in I.J. Inclusion of trials: up to April 24, 2024. Detailed classification is available via the online supporting information.
mRNA-enabled cellular therapy is on the rapid rise, with just a handful of trials breaching into phases II and II/III (Fig. 7, c1). The indications are overwhelming towards oncology (95%) with some application on infectious diseases (Fig. 7, c2). The leading application in oncology is mRNA-enabled antigen-loaded dendritic cell (DC) vaccines, followed by chimeric antigen receptor (CAR)-enabled T cell and NK cell therapies and mRNA-enabled antigen-loaded monocyte vaccines (Fig. 7, c3). Major targets in oncology are led by brain cancer (~30%), followed by melanoma, leukemia, prostate cancer, personalized cancer therapies, and others (Fig. 7, c4).
On the opposite spectrum, the development of siRNA therapies has higher clinical trial completion percentages compared to later-emerging mRNA trials, although the total volume of trials is fewer than mRNA trials (Fig. 7, d1). Clinical applications are led by genetic disorders (~40%), followed by oncology (~17%), eye, liver, and cardiovascular diseases, and others (Fig. 7, d2). Leading siRNA applications in genetic disorders include ATTR amyloidosis (~30%), primary hyperoxaluria, acute intermittent porphyria, hemophilia, myotonic dystrophy, and others (Fig. 7, d3). This is followed by cardiovascular diseases (CVD) and metabolic syndrome, with over 65% of the siRNA trials looking at hypercholesterolemia (exemplified by the approval of Inclisiran in 2021) and atherosclerotic diseases, followed by hypertension, hyper-lipidemia, and thrombosis (Fig. 7, d4). As for oncology, siRNA is most studied for treating solid tumors, which came up on top ~33% of the trials, followed by liver, pancreatic, and skin cancer, and others (Fig. 7, d5). The siRNA-targeted oncogenes are summarized in Table S3. In sum, these pioneering clinical trials chart a transformative course for mRNA and siRNA therapeutics, revealing a horizon rich with potential for addressing some of the most pressing medical challenges of our time.
7. Challenges and future perspectives
Compared to traditional medicinal chemistry-driven drug discovery and development approaches for small molecular drugs, the development of RNA therapeutics, or genetic medicine in general, is often driven by gene sequencing results (e.g., mRNA vaccines or siRNA silencing), protein sequencing results (e.g., for protein replacement therapy), or function-driven protein engineering designs (e.g., CARs), followed by coding such therapeutic blueprints into the RNA sequences for in vivo expression (as mRNA) or silencing (as siRNA). These approaches led to a turnaround in the design and synthesis of RNA molecules for therapeutics compared to the development of new small molecule drugs through traditional chemistry due to the ability to rapidly synthesize RNA once the target sequence is known. More importantly, regardless of the RNA sequence, all could be delivered using LNP platforms to enable clinical translation. Still, significant challenges remained in improving the precision and efficiency of RNA delivery. As precision medicine continues to evolve, the FDA’s validation of RNA-based therapeutics stands as a testament to the burgeoning potential of RNA-enabled treatment strategies. The requirement for precision RNA delivery has been significantly elevated from intramuscular mRNA vaccine injection to CRISPR-based gene editing, demanding the highest level of cell-specific RNA delivery. This precision delivery of RNA therapeutics is contingent on successfully navigating the complex in vivo environment. As both RNA molecules and lipids are inherently unstable and biodegradable under physiological conditions due to lipase or other enzymes, such as the ubiquitous presence of ribonucleases, delivering RNAs to biologically harsh environments, e.g., GI tract, remained highly challenging. Moreover, even when cell-specific RNA delivery is achieved, the efficiency with which these formulations can escape the endolysosomal trafficking once inside the cells is currently limited to approximately 2%, posing another significant barrier for delivering therapeutic RNA to their site of action. Additionally, failing to achieve precision delivery leads to off-target effects within the circulatory system and unintended toxicities. When delivering siRNAs, the off-target effect compounds over a prolonged period, furthering the problem [244,245].
For protein displacement mRNA therapies, e.g., human EPO, the current state of delivering hEPO-encoding mRNA via LNPs shows a duration of expression that has hardly reached beyond 24+ hours [246], a stark contrast to siRNA therapies, which can sustain effects for months. Chemical modification of lipid headgroups (e.g., Lipid 29) was shown to significantly increase the serum EPO concentration for ~3 days [18]. More recently, it was found that codon optimization for attenuating ribosome load plays a critical role in limiting translation-dependent mRNA decay, leading to an overall improved mRNA translation [247]. To address RNA’s inherent instability, advances in chemical modifications have emerged as a pivotal strategy to bolster RNA stability and mitigate immune responses to exogenous mRNA [226]. Modifications like N6-methyladenosine (m6A) and pseudouridine (Ψ) not only fortify the RNA structure but also refine its interaction with cellular machinery, enhancing translation efficacy as seen in mRNA vaccines like Comirnaty® and Spikevax® [248–250]. The logistical demands of RNA therapies, particularly mRNA, which necessitate sub-zero storage conditions, underscore the need for innovation in formulation stability. This necessity was highlighted during the rapid deployment of COVID-19 vaccines and remains an area ripe for improvement. The most recent development by Moderna’s next-generation mRNA-1283 COVID-19 vaccine can now store at 2–5 °C, highlighting another aspect of the importance of LNP formulation development for ensuring RNA stabilities.
The loading capacity of LNPs is another interesting aspect for future development. While most lipidoids were developed for siRNA delivery, delivering mRNA has been challenging due to the sharp increase in nucleotides embedded in mRNA. It is known that payload distribution and RNA loading capacity critically affect LNP formulation’s physicochemical properties and in vivo PK/PD [103]. Developing LNP platforms capable of delivering large proteins, e.g., dystrophin for Duchenne muscular dystrophy, remains challenging. Although a CRISPR-based micro-dystrophin editing approach has been demonstrated for treating the Duchenne muscular dystrophy (DMD) mouse model, the editing effect remained limited by the transient expression of mRNA due to the limitations mentioned above [251].
Immunogenicity adds another layer of complexity to RNA therapy. For example, heterocyclic lipids are known to stimulate immune responses [252,253], which may be favorable when developing immunogenic mRNA vaccines for cancer or infectious diseases. However, when developing tolerogenic mRNA vaccines, the opposite is required, which has been demonstrated by formulating m1Ψ modified mRNA [16] and the incorporation of Dexamethasone lipid-conjugated prodrugs in LNP formulations [254]. More interestingly, the sensory mechanism of endosomal damage caused by the endosomal escape of LNPs is linked to LNP-associated inflammation, leaving additional challenges for noninflammatory RNA delivery [255].
Recent clinical developments reflect the complexity of translating preclinical successes into safe and efficacious human therapies. While liver RNA delivery via LNPs has been widely reported and is considered relatively mature in preclinical mouse models, Verve Therapeutics halted their phase I clinical trial (Heart-1) in April 2024 due to safety and tolerability concerns, which involves their liver targeting CRISPR-LNPs VERVE-101 targets PCSK9 gene for lowering cholesterol [256]. Alternative LNP VERVE-102 is proposed to replace the original ionizable lipid in the VERVE-101 formulation. As the FDA mandates a 14-year followup period for patients who participated in base-editing therapies and trials, this represents one of the highest safety standards requirements for RNA therapeutics and echoes the need for developing organ- and cell-specific LNPs that not only perform and are compatible with preclinical models but, more importantly, to be safe for patient use. Yet, little is known, with more to be investigated to bridge the gap between preclinical and clinical studies of LNPs.
Looking ahead, DNA/RNA barcoding technologies emerge as vital tools in refining LNP formulation discovery. They enable the dissection of biodistribution at a cellular level, fostering efficient preclinical evaluations. The established efficacy of organ-specific LNPs in animal models paves the way for broader applications. Yet, extending cell-specific delivery to a wider array of cell types remains an uncharted and promising frontier for precision medicine. As many of the genome editing targets of LNPs are mostly different from the traditional liposomal biodistribution targets, discovering novel tropism-specific lipids and LNPs remains an up-and-rising challenge with the ever-increasing need for precision RNA therapeutics. In summary, the development of RNA therapeutics stands at a juncture of remarkable potential, where the depth of lipid and RNA engineering converges with the breadth of delivery platforms to chart the course for the next generation of precision RNA medicine.
Supplementary Material
Acknowledgment
This work was partly funded by grants from the Research Foundation for the State University of New York at Binghamton faculty startup fund (910252-69), The SUNY Binghamton Transdisciplinary Areas of Excellence Seed Grant (RF1182424), and the Foundation to Eradicate Duchenne (99387).
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Declaration of generative AI and AI-assisted technologies in the writing process
The author used Microsoft Bing and Grammarly to improve the manuscript’s readability. After using this tool, the authors reviewed and edited the content as needed and took full responsibility for the publication’s content.
CRediT authorship contribution statement
Pu-Sheng Wei: Writing – original draft, Data curation. Nagasri Thota: Writing – original draft, Data curation. Greshma John: Formal analysis, Data curation. Evelyn Chang: Formal analysis, Data curation. Sunjae Lee: Formal analysis, Data curation. Yuanjun Wang: Formal analysis, Data curation. Zitao Ma: Formal analysis, Data curation. Yu-Hsuan Tsai: Writing – review & editing, Formal analysis, Data curation. Kuo-Ching Mei: Writing – review & editing, Writing – original draft, Supervision, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jconrel.2024.08.030.
Data availability
Data will be made available on request.
References
- [1].Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC, Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine, New England Journal of Medicine 383 (2020) 2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T, Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine, New England Journal of Medicine 384 (2020) 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Paramasivam P, Franke C, Stöter M, H00F6ijer A, Bartesaghi S, Sabirsh A, Lindfors L, Arteta MY, Dahlén A, Bak A, Andersson S, Kalaidzidis Y, Bickle M, Zerial M, Endosomal escape of delivered mRNA from endosomal recycling tubules visualized at the nanoscale, J. Cell Biol 221 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chatterjee S, Kon E, Sharma P, Peer D, Endosomal escape: a bottleneck for LNP-mediated therapeutics, Proc. Natl. Acad. Sci 121 (2024) e2307800120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yang T, Li C, Wang X, Zhao D, Zhang M, Cao H, Liang Z, Xiao H, Liang X-J, Weng Y, Huang Y, Efficient hepatic delivery and protein expression enabled by optimized mRNA and ionizable lipid nanoparticle, Bioactive Materials 5 (2020) 1053–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Golubovskaya V, Sienkiewicz J, Sun J, Huang Y, Hu L, Zhou H, Harto H, Xu S, Berahovich R, Bodmer W, Wu L, mRNA-lipid nanoparticle (LNP) delivery of humanized EpCAM-CD3 bispecific antibody significantly blocks colorectal Cancer tumor growth, Cancers 15 (2023) 2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schlake T, Thess A, Thran M, Jordan I, mRNA as novel technology for passive immunotherapy, Cell. Mol. Life Sci 76 (2019) 301–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Billingsley MM, Singh N, Ravikumar P, Zhang R, June CH, Mitchell MJ, Ionizable lipid nanoparticle-mediated mRNA delivery for human CAR T cell engineering, Nano Lett. 20 (2020) 1578–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Miller JB, Zhang S, Kos P, Xiong H, Zhou K, Perelman SS, Zhu H, Siegwart DJ, Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA, Angew. Chem. Int. Ed. Engl 56 (2017) 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wei T, Cheng Q, Min YL, Olson EN, Siegwart DJ, Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing, Nat. Commun 11 (2020) 3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Han JP, Kim M, Choi BS, Lee JH, Lee GS, Jeong M, Lee Y, Kim EA, Oh HK, Go N, Lee H, Lee KJ, Kim UG, Lee JY, Kim S, Chang J, Lee H, Song DW, Yeom SC, In vivo delivery of CRISPR-Cas9 using lipid nanoparticles enables antithrombin gene editing for sustainable hemophilia a and B therapy, Sci. Adv 8 (2022) eabj6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].McKay PF, Hu K, Blakney AK, Samnuan K, Brown JC, Penn R, Zhou J, Bouton CR, Rogers P, Polra K, Lin PJC, Barbosa C, Tam YK, Barclay WS, Shattock RJ, Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice, Nat. Commun 11 (2020) 3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Essink B, Chu L, Seger W, Barranco E, Le Cam N, Bennett H, Faughnan V, Pajon R, Paila YD, Bollman B, Wang S, Dooley J, Kalidindi S, Leav B, The safety and immunogenicity of two Zika virus mRNA vaccine candidates in healthy flavivirus baseline seropositive and seronegative adults: the results of two randomised, placebo-controlled, dose-ranging, phase 1 clinical trials, Lancet Infect. Dis 23 (2023) 621–633. [DOI] [PubMed] [Google Scholar]
- [14].Rojas LA, Sethna Z, Soares KC, Olcese C, Pang N, Patterson E, Lihm J, Ceglia N, Guasp P, Chu A, Yu R, Chandra AK, Waters T, Ruan J, Amisaki M, Zebboudj A, Odgerel Z, Payne G, Derhovanessian E, Müller F, Rhee I, Yadav M, Dobrin A, Sadelain M, Łuksza M, Cohen N, Tang L, Basturk O, G ӧnen M, Katz S, Do RK, Epstein AS, Momtaz P, Park W, Sugarman R, Varghese AM, Won E, Desai A, Wei AC, D’Angelica MI, Kingham TP, Mellman I, Merghoub T, Wolchok JD, Sahin U, Türeci Ö, Greenbaum BD, Jarnagin WR, Drebin J, O’Reilly EM, Balachandran VP, Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer, Nature 618 (2023) 144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Xu X, Wang X, Liao YP, Luo L, Xia T, Nel AE, Use of a liver-targeting immune-Tolerogenic mRNA lipid nanoparticle platform to treat Peanut-induced anaphylaxis by single- and multiple-epitope nucleotide sequence delivery, ACS Nano 17 (2023) 4942–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Krienke C, Kolb L, Diken E, Streuber M, Kirchhoff S, Bukur T, Akilli-Oztürk Ö, Kranz LM, Berger H, Petschenka J, Diken M, Kreiter S, Yogev N, Waisman A, Kariké K,Türeci Ö, Sahin U, A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis, Science 371 (2021) 145–153. [DOI] [PubMed] [Google Scholar]
- [17].Rohner E, Yang R, Foo KS, Goedel A, Chien KR, Unlocking the promise of mRNA therapeutics, Nat. Biotechnol 40 (2022) 1586–1600. [DOI] [PubMed] [Google Scholar]
- [18].Cornebise M, Narayanan E, Xia Y, Acosta E, Ci L, Koch H, Milton J, Sabnis S, Salerno T, Benenato KE, Discovery of a novel amino lipid that improves lipid nanoparticle performance through specific interactions with mRNA, Adv. Funct. Mater 32 (2022) 2106727. [Google Scholar]
- [19].Sheridan C, The world’s first CRISPR therapy is approved: who will receive it? Nat. Biotechnol 42 (2024) 3–4. [DOI] [PubMed] [Google Scholar]
- [20].Xue L, Hamilton AG, Zhao G, Xiao Z, El-Mayta R, Han X, Gong N, Xiong X, Xu J, Figueroa-Espada CG, Shepherd SJ, Mukalel AJ, Alameh M-G, Cui J, Wang K, Vaughan AE, Weissman D, Mitchell MJ, High-throughput barcoding of nanoparticles identifies cationic, degradable lipid-like materials for mRNA delivery to the lungs in female preclinical models, Nat. Commun 15 (2024) 1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li B, Manan RS, Liang S-Q, Gordon A, Jiang A, Varley A, Gao G, Langer R, Xue W, Anderson D, Combinatorial design of nanoparticles for pulmonary mRNA delivery and genome editing, Nat. Biotechnol 41 (2023) 1410–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jiang AY, Witten J, Raji IO, Eweje F, MacIsaac C, Meng S, Oladimeji FA, Hu Y, Manan RS, Langer R, Anderson DG, Combinatorial development of nebulized mRNA delivery formulations for the lungs, Nat. Nanotechnol 19 (2023) 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tsuchida CA, Wasko KM, Hamilton JR, Doudna JA, Targeted nonviral delivery of genome editors in vivo, Proc. Natl. Acad. Sci 121 (2024) e2307796121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pratt AJ, MacRae IJ, The RNA-induced silencing complex: a versatile gene-silencing machine*, J. Biol. Chem 284 (2009) 17897–17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y, Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes, Mol. Cell 39 (2010) 292–299. [DOI] [PubMed] [Google Scholar]
- [26].Kobayashi H, Tomari Y, RISC assembly: coordination between small RNAs and Argonaute proteins, Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1859 (2016) 71–81. [DOI] [PubMed] [Google Scholar]
- [27].Birmingham A, Anderson E, Sullivan K, Reynolds A, Boese Q, Leake D, Karpilow J, Khvorova A, A protocol for designing siRNAs with high functionality and specificity, Nat. Protoc 2 (2007) 2068–2078. [DOI] [PubMed] [Google Scholar]
- [28].Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T, Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells, Nature 411 (2001) 494–498. [DOI] [PubMed] [Google Scholar]
- [29].Urits I, Swanson D, Swett MC, Patel A, Berardino K, Amgalan A, Berger AA, Kassem H, Kaye AD, Viswanath O, A review of Patisiran (ONPATTRO®) for the treatment of polyneuropathy in people with hereditary transthyretin amyloidosis, Neurol Ther 9 (2020) 301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Syed YY, Givosiran: a review in acute hepatic Porphyria, Drugs 81 (2021) 841–848. [DOI] [PubMed] [Google Scholar]
- [31].Garrelfs SF, Frishberg Y, Hulton SA, Koren MJ, O’Riordan WD, Cochat P, Deschênes G, Shasha-Lavsky H, Saland JM, Van’t Hoff WG, Fuster DG, Magen D, Moochhala SH, Schalk G, Simkova E, Groothoff JW, Sas DJ, Meliambro KA, Lu J, Sweetser MT, Garg PP, Vaishnaw AK, Gansner JM, McGregor TL, Lieske JC, Lumasiran, an RNAi therapeutic for primary Hyperoxaluria type 1, N. Engl. J. Med 384 (2021) 1216–1226. [DOI] [PubMed] [Google Scholar]
- [32].Raal FJ, Kallend D, Ray KK, Turner T, Koenig W, Wright RS, Wijngaard PLJ, Curcio D, Jaros MJ, Leiter LA, Kastelein JJP, Inclisiran for the treatment of heterozygous familial hypercholesterolemia, New England Journal of Medicine 382 (2020) 1520–1530. [DOI] [PubMed] [Google Scholar]
- [33].Habtemariam BA, Karsten V, Attarwala H, Goel V, Melch M, Clausen VA, Garg P, Vaishnaw AK, Sweetser MT, Robbie GJ, Vest J, Single-dose pharmacokinetics and pharmacodynamics of transthyretin targeting N-acetylgalactosamine-small interfering ribonucleic acid conjugate, Vutrisiran, in healthy subjects, Clin. Pharmacol. Ther 109 (2021) 372–382. [DOI] [PubMed] [Google Scholar]
- [34].Jensen KK, Tadin-Strapps M, Wang S.-p., Hubert J, Kan Y, Ma Y, McLaren DG, Previs SF, Herath KB, Mahsut A, Liaw A, Wang S, Stout SJ, Keohan C, Forrest G, Coelho D, Yendluri S, Williams S, Koser M, Bartz S, Akinsanya KO, Pinto S, Dose-dependent effects of siRNA-mediated inhibition of SCAP on PCSK9, LDLR, and plasma lipids in mouse and rhesus monkey[S], J. Lipid Res 57 (2016) 2150–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Padda IS, Mahtani AU, Parmar M, Small Interfering RNA (siRNA) Therapy, in: StatPearls, StatPearls PublishingCopyright © 2023, StatPearls Publishing LLC., Treasure Island (FL) Ineligible Companies. Disclosure: Arun Mahtani Declares no Relevant Financial Relationships with Ineligible Companies, Disclosure: Mayur Parmar declares no relevant financial relationships with ineligible companies, 2023. [Google Scholar]
- [36].Doench JG, Petersen CP, Sharp PA, siRNAs can function as miRNAs, Genes Dev. 17 (2003) 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Fedorov Y, Anderson EM, Birmingham A, Reynolds A, Karpilow J, Robinson K, Leake D, Marshall WS, Khvorova A, Off-target effects by siRNA can induce toxic phenotype, RNA 12 (2006) 1188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ilyinskii PO, Michaud AM, Roy CJ, Rizzo GL, Elkins SL, Capela T, Chowdhury AC, Leung SS, Kishimoto TK, Enhancement of liver-directedtransgene expression at initial and repeat doses of AAV vectors admixed with ImmTOR nanoparticles, science, Advances 7 (2021) eabd0 Ö321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bansal D, Abdulmajeed J, Al-Shamali M, Albayat SSA, Himatt SM, Cyprian FS, Chivese T, Mundodan JMA, Khogali HS, Baaboura R, Kaleeckal AH, Kandy MC, Latif AN, Al-Kuwari MG, Al-Romaihi HE, Al Khal A, Bertollini R, Al-Thani MH, Farag E, Doi SAR, Duration of COVID-19 mRNA vaccine effectiveness against severe disease, Vaccines (Basel) 10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity, Science 337 (2012) 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Höijer I, Emmanouilidou A, Östlund R, van Schendel R, Bozorgpana S, Tijsterman M, Feuk L, Gyllensten U, den Hoed M, Ameur A, CRISPR-Cas9 induces large structural variants at on-target and off-target sites in vivo that segregate across generations, Nat. Commun 13 (2022) 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhang X-H, Tee LY, Wang X-G, Huang Q-S, Yang S-H, Off-target effects in CRISPR/Cas9-mediated genome engineering, Molecular Therapy - Nucleic Acids 4 (2015) e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bartoszewski R, Sikorski AF, Editorial focus: understanding off-target effects as the key to successful RNAi therapy, Cell. Mol. Biol. Lett 24 (2019) 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Xu X, Xia T, Recent advances in site-specific lipid nanoparticles for mRNA delivery, ACS Nanoscience Au 3 (2023) 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kim J, Eygeris Y, Ryals RC, Jozić A, Sahay G, Strategies for non-viral vectors targeting organs beyond the liver, Nat. Nanotechnol 19 (2024) 428–447. [DOI] [PubMed] [Google Scholar]
- [46].Narasipura EA, Fenton OS, Advances in non-viral mRNA delivery to the spleen, biomaterials, Biomaterials Sci 12 (2024) 3027–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sun B, Wu W, Narasipura EA, Ma Y, Yu C, Fenton OS, Song H, Engineering nanoparticle toolkits for mRNA delivery, Adv. Drug Deliv. Rev 200 (2023) 115042. [DOI] [PubMed] [Google Scholar]
- [48].Hou X, Zaks T, Langer R, Dong Y, Lipid nanoparticles for mRNA delivery, Nature Reviews Materials 6 (2021) 1078–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Narasipura EA, VanKeulen-Miller R, Ma Y, Fenton OS, Ongoing clinical trials of nonviral siRNA therapeutics, Bioconjug. Chem 34 (2023) 1177–1197. [DOI] [PubMed] [Google Scholar]
- [50].Al-Ahmady Z, Lozano N, Mei K-C, Al-Jamal WT, Kostarelos K, Engineering thermosensitive liposome-nanoparticle hybrids loaded with doxorubicin for heat-triggered drug release, Int. J. Pharm 514 (2016) 133–141. [DOI] [PubMed] [Google Scholar]
- [51].Nel AE, Mei K-C, Liao Y-P, Liu X, Multifunctional lipid bilayer Nanocarriers for Cancer immunotherapy in heterogeneous tumor microenvironments, combining immunogenic cell death stimuli with immune modulatory drugs, ACS Nano 16 (2022) 5184–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mei K-C, Liao Y-P, Jiang J, Chiang M, Khazaieli M, Liu X, Wang X, Liu Q, Chang CH, Zhang X, Li J, Ji Y, Melano B, Telesca D, Xia T, Meng H, Nel AE, Liposomal delivery of Mitoxantrone and a cholesteryl Indoximod prodrug provides effective chemo-immunotherapy in multiple solid tumors, ACS Nano 14 (2020) 13343–13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Schoenmaker L, Witzigmann D, Kulkarni JA, Verbeke R, Kersten G, Jiskoot W, Crommelin DJA, mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability, Int. J. Pharm 601 (2021) 120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yanez Arteta M, Kjellman T, Bartesaghi S, Wallin S, Wu X, Kvist AJ, Dabkowska A, Székely N, Radulescu A, Bergenholtz J, Lindfors L, Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles, Proc. Natl. Acad. Sci 115 (2018) E3351–E3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Patel S, Ashwanikumar N, Robinson E, Xia Y, Mihai C, Griffith JP, Hou S, Esposito AA, Ketova T, Welsher K, Joyal JL, Almarsson Ö, Sahay G, Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA, Nat. Commun 11 (2020) 983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Huang C-H, Phosphatidylcholine vesicles. Formation and physical characteristics, Biochemistry 8 (1969) 344–352. [DOI] [PubMed] [Google Scholar]
- [57].Gregoriadis G, Ryman BE, Liposomes as carriers of enzymes or drugs: a new approach to the treatment of storage diseases, Biochem. J 124 (1971) 58p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Allison AC, Gregoriadis G, Liposomes as immunological adjuvants, Nature 252 (1974) 252. [DOI] [PubMed] [Google Scholar]
- [59].Gregoriadis G, The carrier potential of liposomes in biology and medicine, N. Engl. J. Med 295 (1976) 765–770. [DOI] [PubMed] [Google Scholar]
- [60].Taniguchi N, On the basic concept of ’Nano-Technology’, proceedings of the international conference on, Prod. Eng (1974) 18–23. [Google Scholar]
- [61].Pagano RE, Huang L, Wey C, Interaction of phospholipid vesicles with cultured mammalian cells, Nature 252 (1974) 166–167. [DOI] [PubMed] [Google Scholar]
- [62].Ostro MJ, Giacomoni D, Lavelle DON, Paxton W, Dray S, Evidence for translation of rabbit globin mRNA after liposomemediated insertion into a human cell line, Nature 274 (1978) 921–923. [DOI] [PubMed] [Google Scholar]
- [63].Dimitriadis GJ, Translation of rabbit globin mRNA introduced by liposomes into mouse lymphocytes, Nature 274 (1978) 923–924. [DOI] [PubMed] [Google Scholar]
- [64].Szoka F Jr., Papahadjopoulos D, Procedure for preparation of liposomes with large internal aqueous space and high capture by reverse-phase evaporation, Proc. Natl. Acad. Sci. U. S. A 75 (1978) 4194–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Szoka F, Jacobson K, Derzko Z, Papahadjopoulos D, Fluorescence Studies on the Mechanism of Liposome-Cell Interactions In Vitro, Biochimica et Biophysica Acta (BBA) - Biomembranes vol. 600 (1980) 1–18. [DOI] [PubMed] [Google Scholar]
- [66].Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, Northrop JP, Ringold GM, Danielsen M, Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure, Proc. Natl. Acad. Sci 84 (1987) 7413–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Malone RW, Felgner PL, Verma IM, Cationic liposome-mediated RNA transfection, Proc. Natl. Acad. Sci. U. S. A 86 (1989) 6077–6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Pagano JS, Vaheri A, Enhancement of infectivity of poliovirus RNA with diethylaminoethyl-dextran (DEAE-D), Arch. Gesamte Virusforsch 17 (1965) 456–464. [DOI] [PubMed] [Google Scholar]
- [69].Graham FL, van der Eb AJ, A new technique for the assay of infectivity of human adenovirus 5 DNA, Virology 52 (1973) 456–467. [DOI] [PubMed] [Google Scholar]
- [70].Li S, Gao X, Son K, Sorgi F, Hofland H, Huang L, DC-Chol lipid system in gene transfer, J. Control. Release 39 (1996) 373–381. [Google Scholar]
- [71].Huang L, LPD nanoparticles for gene delivery, Nat. Biotechnol 17 (1999) 18.9920257 [Google Scholar]
- [72].Middleton P, Caplen N, Gao X, Huang L, Gaya H, Geddes D, Alton E, Nasal application of the cationic liposome DC-Chol:DOPE does not alter ion transport, lung function or bacterial growth, Eur. Respir. J 7 (1994) 442–445. [DOI] [PubMed] [Google Scholar]
- [73].Farhood H, Bottega R, Epand RM, Huang L, Effect of Cationic Cholesterol Derivatives on Gene Transfer and Protein Kinase C Activity, Biochimica et Biophysica Acta (BBA) - Biomembranes 1111 (1992) 239–246. [DOI] [PubMed] [Google Scholar]
- [74].Chu CJ, Dijkstra J, Lai MZ, Hong K, Szoka FC, Efficiency of cytoplasmic delivery by pH-sensitive liposomes to cells in culture, Pharm. Res 7 (1990) 824–834. [DOI] [PubMed] [Google Scholar]
- [75].Ellens H, Bentz J, Szoka FC, pH-induced destabilization of phosphatidylethanolamine-containing liposomes: role of bilayer contact, Biochemistry 23 (1984) 1532–1538. [DOI] [PubMed] [Google Scholar]
- [76].Israelachvili JN, Mitchell DJ, Ninham BW, Theory of self-assembly of hydrocarbon amphiphiles into micelles and bilayers, Journal of the Chemical Society, Faraday Transactions 2: Molecular and Chemical Physics 72 (1976) 1525–1568. [Google Scholar]
- [77].Bailey AL, Cullis PR, Modulation of membrane fusion by asymmetric transbilayer distributions of amino lipids, Biochemistry 33 (1994) 12573–12580. [DOI] [PubMed] [Google Scholar]
- [78].Yu H, Angelova A, Angelov B, Dyett B, Matthews L, Zhang Y, El Mohamad M, Cai X, Valimehr S, Drummond CJ, Zhai J, Real-time pH-dependent self-assembly of Ionisable lipids from COVID-19 vaccines and in Situ nucleic acid complexation, Angew. Chem. Int. Ed 62 (2023) e202304977. [DOI] [PubMed] [Google Scholar]
- [79].Horejs C, From lipids to lipid nanoparticles to mRNA vaccines, Nature Reviews Materials 6 (2021) 1075–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Han X, Zhang H, Butowska K, Swingle KL, Alameh M-G, Weissman D, Mitchell MJ, An ionizable lipid toolbox for RNA delivery, Nat. Commun 12 (2021) 7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Hafez IM, Ansell S, Cullis PR, Tunable pH-sensitive liposomes composed of mixtures of cationic and anionic lipids, Biophys. J 79 (2000) 1438–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Jayaraman M, Ansell SM, Mui BL, Tam YK, Chen J, Du X, Butler D, Eltepu L, Matsuda S, Narayanannair JK, Rajeev KG, Hafez IM, Akinc A, Maier MA, Tracy MA, Cullis PR, Madden TD, Manoharan M, Hope MJ, Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo**, Angew. Chem. Int. Ed 51 (2012) 8529–8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Lewis RNAH, Zhang Y-P, McElhaney RN, Calorimetric and spectroscopic studies of the phase behavior and organization of lipid bilayer model membranes composed of binary mixtures of dimyristoylphosphatidylcholine and dimyristoylphosphatidylglycerol, Biochimica et Biophysica Acta (BBA) - Biomembranes 1668 (2005) 203–214. [DOI] [PubMed] [Google Scholar]
- [84].Lu T, ten Hagen TLM, Inhomogeneous crystal grain formation in DPPC-DSPC based thermosensitive liposomes determines content release kinetics, J. Control. Release 247 (2017) 64–72. [DOI] [PubMed] [Google Scholar]
- [85].Nagle JF, Theory of lipid monolayer and bilayer phase transitions: effect of headgroup interactions, J. Membr. Biol 27 (1976) 233–250. [DOI] [PubMed] [Google Scholar]
- [86].Ellens H, Bentz J, Szoka FC, Destabilization of phosphatidylethanolamine liposomes at the hexagonal phase transition temperature, Biochemistry 25 (1986) 285–294. [DOI] [PubMed] [Google Scholar]
- [87].Rappolt M, Hickel A, Bringezu F, Lohner K, Mechanism of the lamellar/inverse hexagonal phase transition examined by High resolution X-Ray diffraction, Biophys. J 84 (2003) 3111–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Abuchowski A, McCoy JR, Palczuk NC, van Es T, Davis FF, Effect of covalent attachment of polyethylene glycol on immunogenicity and circulating life of bovine liver catalase, J. Biol. Chem 252 (1977) 3582–3586. [PubMed] [Google Scholar]
- [89].Matsumura Y, Maeda H, A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs, Cancer Res. 46 (1986) 6387–6392. [PubMed] [Google Scholar]
- [90].Klibanov AL, Maruyama K, Torchilin VP, Huang L, Amphipathic polyethyleneglycols effectively prolong the circulation time of liposomes, FEBS Lett. 268 (1990) 235–237. [DOI] [PubMed] [Google Scholar]
- [91].Barenholz Y, Doxil® — the first FDA-approved nano-drug: lessons learned, J. Control. Release 160 (2012) 117–134. [DOI] [PubMed] [Google Scholar]
- [92].Bai J, Wang JT-W, Mei K-C, Al-Jamal WT, Al-Jamal KT, Real-time monitoring of magnetic drug targeting using fibered confocal fluorescence microscopy, J. Control. Release 244 (2016) 240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Rubio N, Mei K-C, Klippstein R, Costa PM, Hodgins N, Wang JT-W, Festy F, Abbate V, Hider RC, Chan KLA, Al-Jamal KT, Solvent-free click-Mechanochemistry for the preparation of Cancer cell targeting graphene oxide, ACS Appl. Mater. Interfaces 7 (2015) 18920–18923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Mei K-C, Bai J, Lorrio S, Wang JT-W, Al-Jamal KT, Investigating the effect of tumor vascularization on magnetic targeting in vivo using retrospective design of experiment, Biomaterials 106 (2016) 276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Etter EL, Mei K-C, Nguyen J, Delivering more for less: nanosized, minimal-carrier and pharmacoactive drug delivery systems, Adv. Drug Deliv. Rev 179 (2021) 113994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Dubey RD, Klippstein R, Wang JT, Hodgins N, Mei KC, Sosabowski J, Hider RC, Abbate V, Gupta PN, Al-Jamal KT, Novel hyaluronic acid conjugates for dual nuclear imaging and therapy in CD44-expressing tumors in mice in vivo, Nanotheranostics 1 (2017) 59–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Mei K-C, Costa PM, Kreuzer M, Al-Jamal KT, Interpreting 2D materials bio-Nano interactions: influence of aggregation status, Protein Corona, Cell Culture Media, and Cell Types, Advanced Materials Interfaces 8 (2021) 2100251. [Google Scholar]
- [98].Mei K-C, Ghazaryan A, Teoh EZ, Summers HD, Li Y, Ballesteros B, Piasecka J, Walters A, Hider RC, Mailӓnder V, Al-Jamal KT, Protein-Corona-by-Design in 2D: A Reliable Platform to Decode Bio–Nano Interactions for the Next-Generation Quality-by-Design Nanomedicines, Adv. Mater 30 (2018) 1802732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Liu X, Jiang J, Liao Y-P, Tang I, Zheng E, Qiu W, Lin M, Wang X, Ji Y, Mei K-C, Liu Q, Chang CH, Wainberg ZA, Nel AE, Meng H, Combination chemo-immunotherapy for pancreatic Cancer using the immunogenic effects of an irinotecan Silicasome Nanocarrier plus anti-PD-1, Advanced Science 8 (2021) 2002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Liu X, Jiang J, Chang CH, Liao Y-P, Lodico JJ, Tang I, Zheng E, Qiu W, Lin M, Wang X, Ji Y, Mei K-C, Nel AE, Meng H, Development of facile and versatile platinum drug delivering Silicasome Nanocarriers for efficient pancreatic Cancer chemo-immunotherapy, Small 17 (2021) 2005993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Mui BL, Tam YK, Jayaraman M, Ansell SM, Du X, Tam YYC, Lin PJC, Chen S, Narayanannair JK, Rajeev KG, Manoharan M, Akinc A, Maier MA, Cullis P, Madden TD, Hope MJ, Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles, Molecular Therapy - Nucleic Acids 2 (2013) e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Muramatsu H, Lam K, Bajusz C, Laczkó D, Karikó K, Schreiner P, Martin A, Lutwyche P, Heyes J, Pardi N, Lyophilization provides long-term stability for a lipid nanoparticle-formulated, nucleoside-modified mRNA vaccine, Mol. Ther 30 (2022) 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Li S, Hu Y, Li A, Lin J, Hsieh K, Schneiderman Z, Zhang P, Zhu Y, Qiu C, Kokkoli E, Wang T-H, Mao H-Q, Payload distribution and capacity of mRNA lipid nanoparticles, Nat. Commun 13 (2022) 5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Characterization of cationic liposome-mediated gene transfer in vivo by intravenous administration, Hum. Gene Ther 8 (1997) 1585–1594. [DOI] [PubMed] [Google Scholar]
- [105].Kolašinac R, Kleusch C, Braun T, Merkel R, Csiszár A, Deciphering the functional composition of Fusogenic liposomes, Int. J. Mol. Sci 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kim B-K, Hwang G-B, Seu Y-B, Choi J-S, Jin KS, Doh K-O, DOTAP/DOPE ratio and cell type determine transfection efficiency with DOTAP-liposomes, Biochimica et Biophysica Acta (BBA) - Biomembranes 1848 (2015) 1996–2001. [DOI] [PubMed] [Google Scholar]
- [107].Singer SJ, Nicolson GL, The fluid mosaic model of the structure of cell membranes, Science 175 (1972) 720–731. [DOI] [PubMed] [Google Scholar]
- [108].Cooper RA, Influence of increased membrane cholesterol on membrane fluidity and cell function in human red blood cells, J. Supramol. Struct 8 (1978) 413–430. [DOI] [PubMed] [Google Scholar]
- [109].Redondo-Morata L, Giannotti MI, Sanz F, Influence of cholesterol on the phase transition of lipid bilayers: a temperature-controlled force spectroscopy study, Langmuir 28 (2012) 12851–12860. [DOI] [PubMed] [Google Scholar]
- [110].Regelin AE, Fankhaenel S, Gürtesch L, Prinz C, von Kiedrowski G, Massing U, Biophysical and lipofection studies of DOTAP analogs, Biochimica et Biophysica Acta (BBA) - Biomembranes 1464 (2000) 151–164. [DOI] [PubMed] [Google Scholar]
- [111].Herrera M, Kim J, Eygeris Y, Jozic A, Sahay G, Illuminating endosomal escape of polymorphic lipid nanoparticles that boost mRNA delivery, Biomater. Sci 9 (2021) 4289–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Patel SK, Billingsley MM, Mukalel AJ, Thatte AS, Hamilton AG, Gong N, El-Mayta R, Safford HC, Merolle M, Mitchell MJ, Bile acid-containing lipid nanoparticles enhance extrahepatic mRNA delivery, Theranostics 14 (2024) 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Ellens H, Bentz J, Szoka FC, pH-induced destabilization of phosphatidylethanolamine-containing liposomes: role of bilayer contact, Biochemistry 23 (1984) 1532–1538. [DOI] [PubMed] [Google Scholar]
- [114].Du S, Li W, Zhang Y, Xue Y, Hou X, Yan J, Cheng J, Deng B, McComb DW, Lin J, Zeng H, Cheng X, Irvine DJ, Weiss R, Dong Y, Cholesterol-amino-phosphate (CAP) derived lipid nanoparticles for delivery of self-amplifying RNA and restoration of spermatogenesis in infertile mice, Advanced Science 10 (2023) 2300188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Zhao YC, Zhang L, Feng SS, Hong L, Zheng HL, Chen LL, Zheng XL, Ye YQ, Zhao MD, Wang WX, Zheng CH, Efficient delivery of Notch1 siRNA to SKOV3 cells by cationic cholesterol derivative-based liposome, Int. J. Nanomedicine 11 (2016) 5485–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Patel S, Ashwanikumar N, Robinson E, Xia Y, Mihai C, Griffith JP 3rd, Hou S, Esposito AA, Ketova T, Welsher K, Joyal JL, Almarsson Ö, Sahay G, Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA, Nat. Commun 11 (2020) 983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Sun D, Lu Z-R, Structure and function of cationic and Ionizable lipids for nucleic acid delivery, Pharm. Res 40 (2023) 27–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Heyes J, Palmer L, Bremner K, MacLachlan I, Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids, J. Control. Release 107 (2005) 276–287. [DOI] [PubMed] [Google Scholar]
- [119].Coelho T, Adams D, Silva A, Lozeron P, Hawkins PN, Mant T, Perez J, Chiesa J, Warrington S, Tranter E, Munisamy M, Falzone R, Harrop J, Cehelsky J, Bettencourt BR, Geissler M, Butler JS, Sehgal A, Meyers RE, Chen Q, Borland T, Hutabarat RM, Clausen VA, Alvarez R, Fitzgerald K, Gamba-Vitalo C, Nochur SV, Vaishnaw AK, Sah DWY, Gollob JA, Suhr OB, Safety and efficacy of RNAi therapy for transthyretin amyloidosis, New England Journal of Medicine 369 (2013) 819–829. [DOI] [PubMed] [Google Scholar]
- [120].Maier MA, Jayaraman M, Matsuda S, Liu J, Barros S, Querbes W, Tam YK, Ansell SM, Kumar V, Qin J, Zhang X, Wang Q, Panesar S, Hutabarat R, Carioto M, Hettinger J, Kandasamy P, Butler D, Rajeev KG, Pang B, Charisse K, Fitzgerald K, Mui BL, Du X, Cullis P, Madden TD, Hope MJ, Manoharan M, Akinc A, Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics, Mol. Ther 21 (2013) 1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Kenjo E, Hozumi H, Makita Y, Iwabuchi KA, Fujimoto N, Matsumoto S, Kimura M, Amano Y, Ifuku M, Naoe Y, Inukai N, Hotta A, Low immunogenicity of LNP allows repeated administrations of CRISPR-Cas9 mRNA into skeletal muscle in mice, Nat. Commun 12 (2021) 7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Tanaka H, Sakurai Y, Anindita J, Akita H, Development of lipid-like materials for RNA delivery based on intracellular environment-responsive membrane destabilization and spontaneous collapse, Adv. Drug Deliv. Rev 154–155 (2020) 210–226. [DOI] [PubMed] [Google Scholar]
- [123].Sabnis S, Kumarasinghe ES, Salerno T, Mihai C, Ketova T, Senn JJ, Lynn A, Bulychev A, McFadyen I, Chan J, Almarsson Ö, Stanton MG, Benenato KE, A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human Primates, Mol. Ther 26 (2018) 1509–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Conway A, Mendel M, Kim K, McGovern K, Boyko A, Zhang L, Miller JC, DeKelver RC, Paschon DE, Mui BL, Lin PJC, Tam YK, Barbosa C, Redelmeier T, Holmes MC, Lee G, Non-viral delivery of zinc finger nuclease mRNA enables highly efficient in vivo genome editing of multiple therapeutic gene targets, Mol. Ther 27 (2019) 866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Li W, Zhang X, Zhang C, Yan J, Hou X, Du S, Zeng C, Zhao W, Deng B, McComb DW, Zhang Y, Kang DD, Li J, Carson WE, Dong Y, Biomimetic nanoparticles deliver mRNAs encoding costimulatory receptors and enhance T cell mediated cancer immunotherapy, Nat. Commun 12 (2021) 7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Suzuki Y, Hyodo K, Suzuki T, Tanaka Y, Kikuchi H, Ishihara H, Biodegradable lipid nanoparticles induce a prolonged RNA interference-mediated protein knockdown and show rapid hepatic clearance in mice and nonhuman primates, Int. J. Pharm 519 (2017) 34–43. [DOI] [PubMed] [Google Scholar]
- [127].Rhym LH, Manan RS, Koller A, Stephanie G, Anderson DG, Peptide-encoding mRNA barcodes for the high-throughput in vivo screening of libraries of lipid nanoparticles for mRNA delivery, Nat. Biomed. Eng 7 (2023) 901–910. [DOI] [PubMed] [Google Scholar]
- [128].Gillmore JD, Gane E, Taubel J, Kao J, Fontana M, Maitland ML, Seitzer J, O’Connell D, Walsh KR, Wood K, Phillips J, Xu Y, Amaral A, Boyd AP, Cehelsky JE, McKee MD, Schiermeier A, Harari O, Murphy A, Kyratsous CA, Zambrowicz B, Soltys R, Gutstein DE, Leonard J, Sepp-Lorenzino L, Lebwohl D, CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis, New England Journal of Medicine 385 (2021) 493–502. [DOI] [PubMed] [Google Scholar]
- [129].Finn JD, Smith AR, Patel MC, Shaw L, Youniss MR, van Heteren J, Dirstine T, Ciullo C, Lescarbeau R, Seitzer J, Shah RR, Shah A, Ling D, Growe J, Pink M, Rohde E, Wood KM, Salomon WE, Harrington WF, Dombrowski C, Strapps WR, Chang Y, Morrissey DV, A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing, Cell Rep. 22 (2018) 2227–2235. [DOI] [PubMed] [Google Scholar]
- [130].Han X, Xu J, Xu Y, Alameh M-G, Xue L, Gong N, El-Mayta R, Palanki R, Warzecha CC, Zhao G, Vaughan AE, Wilson JM, Weissman D, Mitchell MJ, In situ combinatorial synthesis of degradable branched lipidoids for systemic delivery of mRNA therapeutics and gene editors, Nat. Commun 15 (2024) 1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Zhao X, Chen J, Qiu M, Li Y, Glass Z, Xu Q, Imidazole-based synthetic Lipidoids for in vivo mRNA delivery into primary T lymphocytes, Angew. Chem. Int. Ed 59 (2020) 20083–20089. [DOI] [PubMed] [Google Scholar]
- [132].Wang M, Zuris JA, Meng F, Rees H, Sun S, Deng P, Han Y, Gao X, Pouli D, Wu Q, Georgakoudi I, Liu DR, Xu Q, Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles, Proc. Natl. Acad. Sci 113 (2016) 2868–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Li Y, Bolinger J, Yu Y, Glass Z, Shi N, Yang L, Wang M, Xu Q, Intracellular delivery and biodistribution study of CRISPR/Cas9 ribonucleoprotein loaded bioreducible lipidoid nanoparticles, biomaterials, Science 7 (2019) 596–606. [DOI] [PubMed] [Google Scholar]
- [134].Li Y, Ye Z, Yang H, Xu Q, Tailoring combinatorial lipid nanoparticles for intracellular delivery of nucleic acids, proteins, and drugs, Acta Pharm. Sin. B 12 (2022) 2624–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Akinc A, Lynn DM, Anderson DG, Langer R, Parallel synthesis and biophysical characterization of a degradable polymer library for gene delivery, J. Am. Chem. Soc 125 (2003) 5316–5323. [DOI] [PubMed] [Google Scholar]
- [136].Lynn DM, Anderson DG, Putnam D, Langer R, Accelerated discovery of synthetic transfection vectors: parallel synthesis and screening of a degradable polymer library, J. Am. Chem. Soc 123 (2001) 8155–8156. [DOI] [PubMed] [Google Scholar]
- [137].Anderson DG, Lynn DM, Langer R, Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery, Angew. Chem. Int. Ed 42 (2003) 3153–3158. [DOI] [PubMed] [Google Scholar]
- [138].Lynn DM, Langer R, Degradable poly(β-amino esters): synthesis, characterization, and self-assembly with plasmid DNA, J. Am. Chem. Soc 122 (2000) 10761–10768. [Google Scholar]
- [139].Lynn DM, Amiji MM, Langer R, pH-responsive polymer microspheres: rapid release of encapsulated material within the range of intracellular pH, Angew. Chem. Int. Ed 40 (2001) 1707–1710. [PubMed] [Google Scholar]
- [140].Afanasyev OI, Kuchuk E, Usanov DL, Chusov D, Reductive amination in the synthesis of pharmaceuticals, Chem. Rev 119 (2019) 11857–11911. [DOI] [PubMed] [Google Scholar]
- [141].Whitehead KA, Sahay G, Li GZ, Love KT, Alabi CA, Ma M, Zurenko C, Querbes W, Langer RS, Anderson DG, Synergistic silencing: combinations of lipid-like materials for efficacious siRNA delivery, Mol. Ther 19 (2011) 1688–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, Borodovsky A, Borland T, Constien R, de Fougerolles A, Dorkin JR, Narayanannair Jayaprakash K, Jayaraman M, John M, Koteliansky V, Manoharan M, Nechev L, Qin J, Racie T, Raitcheva D, Rajeev KG, Sah DWY, Soutschek J, Toudjarska I, Vornlocher H-P, Zimmermann TS, Langer R, Anderson DG, A combinatorial library of lipid-like materials for delivery of RNAi therapeutics, Nat. Biotechnol 26 (2008) 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Akinc A, Goldberg M, Qin J, Dorkin JR, Gamba-Vitalo C, Maier M, Jayaprakash KN, Jayaraman M, Rajeev KG, Manoharan M, Koteliansky V, Röhl I, Leshchiner ES, Langer R, Anderson DG, Development of Lipidoid–siRNA formulations for systemic delivery to the liver, Mol. Ther 17 (2009) 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, Frank-Kamenetsky M, Yip KN, Alvarez R, Sah DWY, de Fougerolles A, Fitzgerald K, Koteliansky V, Akinc A, Langer R, Anderson DG, Lipid-like materials for low-dose, in vivo gene silencing, Proc. Natl. Acad. Sci 107 (2010) 1864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Kim M, Jeong M, Hur S, Cho Y, Park J, Jung H, Seo Y, Woo HA, Nam KT, Lee K, Lee H, Engineered ionizable lipid nanoparticles for targeted delivery of RNA therapeutics into different types of cells in the liver, Sci. Adv 7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Riley RS, Kashyap MV, Billingsley MM, White B, Alameh M-G, Bose SK, Zoltick PW, Li H, Zhang R, Cheng AY, Weissman D, Peranteau WH, Mitchell MJ, Ionizable lipid nanoparticles for in utero mRNA delivery, Science, Advances 7 (2021) eaba1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Han X, Alameh M-G, Butowska K, Knox JJ, Lundgreen K, Ghattas M, Gong N, Xue L, Xu Y, Lavertu M, Bates P, Xu J, Nie G, Zhong Y, Weissman D, Mitchell MJ, Adjuvant lipidoid-substituted lipid nanoparticles augment the immunogenicity of SARS-CoV-2 mRNA vaccines, Nat. Nanotechnol 18 (2023) 1105–1114. [DOI] [PubMed] [Google Scholar]
- [148].Tilstra G, Couture-Senécal J, Lau YMA, Manning AM, Wong DSM, Janaeska WW, Wuraola TA, Pang J, Khan OF, Iterative Design of Ionizable Lipids for intramuscular mRNA delivery, J. Am. Chem. Soc 145 (2023) 2294–2304. [DOI] [PubMed] [Google Scholar]
- [149].Ni H, Hatit MZC, Zhao K, Loughrey D, Lokugamage MP, Peck HE, Cid AD, Muralidharan A, Kim Y, Santangelo PJ, Dahlman JE, Piperazine-derived lipid nanoparticles deliver mRNA to immune cells in vivo, Nat. Commun 13 (2022) 4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Mukalel AJ, Hamilton AG, Billingsley MM, Li J, Thatte AS, Han X, Safford HC, Padilla MS, Papp T, Parhiz H, Weissman D, Mitchell MJ, Oxidized mRNA Lipid Nanoparticles for In Situ Chimeric Antigen Receptor Monocyte Engineering, Advanced Functional Materials (2024) 2312038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, Fenton OS, Zhang Y, Olejnik KT, Yesilyurt V, Chen D, Barros S, Klebanov B, Novobrantseva T, Langer R, Anderson DG, Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity, Nat. Commun 5 (2014) 4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, Bogorad RL, Yin H, Chen Y, Vegas AJ, Alabi CA, Sahay G, Olejnik KT, Wang W, Schroeder A, Lytton-Jean AKR, Siegwart DJ, Akinc A, Barnes C, Barros SA, Carioto M, Fitzgerald K, Hettinger J, Kumar V, Novobrantseva TI, Qin J, Querbes W, Koteliansky V, Langer R, Anderson DG, Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates, Proc. Natl. Acad. Sci 111 (2014) 3955–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Fenton OS, Kauffman KJ, McClellan RL, Appel EA, Dorkin JR, Tibbitt MW, Heartlein MW, DeRosa F, Langer R, Anderson DG, Bioinspired alkenyl amino alcohol Ionizable lipid materials for highly potent in vivo mRNA delivery, Adv. Mater 28 (2016) 2939–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Fenton OS, Kauffman KJ, Kaczmarek JC, McClellan RL, Jhunjhunwala S, Tibbitt MW, Zeng MD, Appel EA, Dorkin JR, Mir FF, Yang JH, Oberli MA, Heartlein MW, DeRosa F, Langer R, Anderson DG, Synthesis and biological evaluation of Ionizable lipid materials for the in vivo delivery of messenger RNA to B lymphocytes, Adv. Mater 29 (2017) 1606944. [DOI] [PubMed] [Google Scholar]
- [155].Zhang X, Zhao W, Nguyen GN, Zhang C, Zeng C, Yan J, Du S, Hou X, Li W, Jiang J, Deng B, McComb DW, Dorkin R, Shah A, Barrera L, Gregoire F, Singh M, Chen D, Sabatino DE, Dong Y, Functionalized lipid-like nanoparticles for in vivo mRNA delivery and base editing, science, Advances 6 (2020) eabc2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Zhang D, Wang G, Yu X, Wei T, Farbiak L, Johnson LT, Taylor AM, Xu J, Hong Y, Zhu H, Siegwart DJ, Enhancing CRISPR/Cas gene editing through modulating cellular mechanical properties for cancer therapy, Nat. Nanotechnol 17 (2022) 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Dahlman JE, Barnes C, Khan OF, Thiriot A, Jhunjunwala S, Shaw TE, Xing Y, Sager HB, Sahay G, Speciner L, Bader A, Bogorad RL, Yin H, Racie T, Dong Y, Jiang S, Seedorf D, Dave A, Singh Sandhu K, Webber MJ, Novobrantseva T, Ruda VM, Lytton-Jean AKR, Levins CG, Kalish B, Mudge DK, Perez M, Abezgauz L, Dutta P, Smith L, Charisse K, Kieran MW, Fitzgerald K, Nahrendorf M, Danino D, Tuder RM, von Andrian UH, Akinc A, Panigrahy D, Schroeder A, Koteliansky V, Langer R, Anderson DG, In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight, Nat. Nanotechnol 9 (2014) 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Yu X, Liu S, Cheng Q, Wei T, Lee S, Zhang D, Siegwart DJ, Lipid-modified aminoglycosides for mRNA delivery to the liver, Adv. Healthc. Mater 9 (2020) 1901487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Gabizon A, Goren D, Fuks Z, Meshorer A, Barenholz Y, Superior therapeutic activity of liposome-associated adriamycin in a murine metastatic tumour model, Br. J. Cancer 51 (1985) 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Wilhelm S, Tavares AJ, Dai Q, Ohta S, Audet J, Dvorak HF, Chan WCW, Analysis of nanoparticle delivery to tumours, Nat. Rev. Mater 1 (2016) 16014. [Google Scholar]
- [161].Liu D, Mori A, Huang L, Role of liposome size and RES blockade in controlling biodistribution and tumor uptake of GM1-containing liposomes, Biochimica et Biophysica Acta (BBA) - Biomembranes 1104 (1992) 95–101. [DOI] [PubMed] [Google Scholar]
- [162].Abraham I, Goundalkar A, Mezei M, Effect of liposomal surface charge on the pharmacokinetics of an encapsulated model compound, Biopharm. Drug Dispos 5 (1984) 387–398. [DOI] [PubMed] [Google Scholar]
- [163].Zhu N, Liggitt D, Liu Y, Debs R, Systemic gene expression after intravenous DNA delivery into adult mice, Science 261 (1993) 209–211. [DOI] [PubMed] [Google Scholar]
- [164].Tan Y, Liu F, Li Z, Li S, Huang L, Sequential injection of cationic liposome and plasmid DNA effectively transfects the lung with minimal inflammatory toxicity, Mol. Ther 3 (2001) 673–682. [DOI] [PubMed] [Google Scholar]
- [165].Sakurai F, Nishioka T, Yamashita F, Takakura Y, Hashida M, Effects of erythrocytes and serum proteins on lung accumulation of lipoplexes containing cholesterol or DOPE as a helper lipid in the single-pass rat lung perfusion system, Eur. J. Pharm. Biopharm 52 (2001) 165–172. [DOI] [PubMed] [Google Scholar]
- [166].Gregory Gregoriadis E, Neerunjun D, Homing of liposomes to target cells, Biochem. Biophys. Res. Commun 65 (1975) 537–544. [DOI] [PubMed] [Google Scholar]
- [167].Magee WE, Cronenberger JH, Thor DE, Marked stimulation of lymphocyte-mediated attack on tumor cells by target-directed liposomes containing immune RNA, Cancer Res. 38 (1978) 1173–1176. [PubMed] [Google Scholar]
- [168].Huang A, Huang L, Kennel SJ, Monoclonal antibody covalently coupled with fatty acid. A reagent for in vitro liposome targeting, J Biol Chem 255 (1980) 8015–8018. [PubMed] [Google Scholar]
- [169].Torchilin VP, Khaw BA, Smirnov VN, Haber E, Preservation of antimyosin antibody activity after covalent coupling to liposomes, Biochem. Biophys. Res. Commun 89 (1979) 1114–1119. [DOI] [PubMed] [Google Scholar]
- [170].Holmberg E, Maruyama K, Kennel S, Klibanov A, Torchilin V, Ryan U, Huang L, Target-specific binding of Immunoliposomes in vivo, J. Liposome Res 1 (1990) 393–406. [Google Scholar]
- [171].Senior M, Cancer-targeting antibody–drug conjugates drive dealmaking frenzy, Nat. Biotechnol 42 (2024) 362–366. [DOI] [PubMed] [Google Scholar]
- [172].Wang X, Liu S, Sun Y, Yu X, Lee SM, Cheng Q, Wei T, Gong J, Robinson J, Zhang D, Lian X, Basak P, Siegwart DJ, Preparation of selective organ-targeting (SORT) lipid nanoparticles (LNPs) using multiple technical methods for tissue-specific mRNA delivery, Nat. Protoc 18 (2023) 265–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Cheng Q, Wei T, Farbiak L, Johnson LT, Dilliard SA, Siegwart DJ, Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing, Nat. Nanotechnol 15 (2020) 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].LoPresti ST, Arral ML, Chaudhary N, Whitehead KA, The replacement of helper lipids with charged alternatives in lipid nanoparticles facilitates targeted mRNA delivery to the spleen and lungs, J. Control. Release 345 (2022) 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Massaro M, Wu S, Baudo G, Liu H, Collum S, Lee H, Stigliano C, Segura-Ibarra V, Karmouty-Quintana H, Blanco E, Lipid nanoparticle-mediated mRNA delivery in lung fibrosis, Eur. J. Pharm. Sci 183 (2023) 106370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Urits I, Swanson D, Swett MC, Patel A, Berardino K, Amgalan A, Berger AA, Kassem H, Kaye AD, Viswanath O, A review of Patisiran (ONPATTRO®) for the treatment of polyneuropathy in people with hereditary transthyretin amyloidosis, Neurology and Therapy 9 (2020) 301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Swingle KL, Safford HC, Geisler HC, Hamilton AG, Thatte AS, Billingsley MM, Joseph RA, Mrksich K, Padilla MS, Ghalsasi AA, Alameh M-G, Weissman D, Mitchell MJ, Ionizable lipid nanoparticles for in vivo mRNA delivery to the placenta during pregnancy, J. Am. Chem. Soc 145 (2023) 4691–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Huang C, Duan X, Wang J, Tian Q, Ren Y, Chen K, Zhang Z, Li Y, Feng Y, Zhong K, Wang Y, Zhou L, Guo G, Song X, Tong A, Lipid nanoparticle delivery system for mRNA encoding B7H3-redirected bispecific antibody displays potent antitumor effects on malignant tumors, Adv Sci (Weinh) 10 (2023) e2205532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Zhang R, El-Mayta R, Murdoch TJ, Warzecha CC, Billingsley MM, Shepherd SJ, Gong N, Wang L, Wilson JM, Lee D, Mitchell MJ, Helper lipid structure influences protein adsorption and delivery of lipid nanoparticles to spleen and liver, biomaterials, Science 9 (2021) 1449–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Gao K, Li J, Song H, Han H, Wang Y, Yin B, Farmer DL, Murthy N, Wang A, In utero delivery of mRNA to the heart, diaphragm and muscle with lipid nanoparticles, Bioactive Materials 25 (2023) 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Pardi N, Tuyishime S, Muramatsu H, Kariko K, Mui BL, Tam YK, Madden TD, Hope MJ, Weissman D, Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes, J. Control. Release 217 (2015) 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Melamed JR, Yerneni SS, Arral ML, LoPresti ST, Chaudhary N, Sehrawat A, Muramatsu H, Alameh M-G, Pardi N, Weissman D, Gittes GK, Whitehead KA, Ionizable lipid nanoparticles deliver mRNA to pancreatic β cells via macrophage-mediated gene transfer, science, Advances 9 (2023) eade1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Farrell KB, Karpeisky A, Thamm DH, Zinnen S, Bisphosphonate conjugation for bone specific drug targeting, Bone Reports 9 (2018) 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Wang G, Mostafa NZ, Incani V, Kucharski C, Uludağ H, Bisphosphonate-decorated lipid nanoparticles designed as drug carriers for bone diseases, J. Biomed. Mater. Res. A 100A (2012) 684–693. [DOI] [PubMed] [Google Scholar]
- [185].Xue L, Gong N, Shepherd SJ, Xiong X, Liao X, Han X, Zhao G, Song C, Huang X, Zhang H, Padilla MS, Qin J, Shi Y, Alameh MG, Pochan DJ, Wang K, Long F, Weissman D, Mitchell MJ, Rational Design of Bisphosphonate Lipid-like Materials for mRNA delivery to the bone microenvironment, J. Am. Chem. Soc 144 (2022) 9926–9937. [DOI] [PubMed] [Google Scholar]
- [186].Lian X, Chatterjee S, Sun Y, Dilliard SA, Moore S, Xiao Y, Bian X, Yamada K, Sung Y-C, Levine RM, Mayberry K, John S, Liu X, Smith C, Johnson LT, Wang X, Zhang CC, Liu DR, Newby GA, Weiss MJ, Yen JS, Siegwart DJ, Bone-marrow-homing lipid nanoparticles for genome editing in diseased and malignant haematopoietic stem cells, Nat. Nanotechnol (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Russell S, Bennett J, Wellman JA, Chung DC, Yu Z-F, Tillman A, Wittes J, Pappas J, Elci O, McCague S, Cross D, Marshall KA, Walshire J, Kehoe TL, Reichert H, Davis M, Raffini L, George LA, Hudson FP, Dingfield L, Zhu X, Haller JA, Sohn EH, Mahajan VB, Pfeifer W, Weckmann M, Johnson C, Gewaily D, Drack A, Stone E, Wachtel K, Simonelli F, Leroy BP, Wright JF, High KA, Maguire AM, Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial, Lancet 390 (2017) 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Patel S, Ryals RC, Weller KK, Pennesi ME, Sahay G, Lipid nanoparticles for delivery of messenger RNA to the back of the eye, J. Control. Release 303 (2019) 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Lv J, Wang H, Cheng X, Chen Y, Wang D, Zhang L, Cao Q, Tang H, Hu S, Gao K, Xun M, Wang J, Wang Z, Zhu B, Cui C, Gao Z, Guo L, Yu S, Jiang L, Yin Y, Zhang J, Chen B, Wang W, Chai R, Chen Z-Y, Li H, Shu Y, AAV1-hOTOF gene therapy for autosomal recessive deafness 9: a single-arm trial, The Lancet 403 (10441) (2024) 2317–2325. [DOI] [PubMed] [Google Scholar]
- [190].Qi J, Tan F, Zhang L, Lu L, Zhang S, Zhai Y, Lu Y, Qian X, Dong W, Zhou Y, Zhang Z, Yang X, Jiang L, Yu C, Liu J, Chen T, Wu L, Tan C, Sun S, Song H, Shu Y, Xu L, Gao X, Li H, Chai R, AAV-mediated gene therapy restores hearing in patients with DFNB9 deafness, Advanced Science 11 (2024) 2306788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Gao X, Tao Y, Lamas V, Huang M, Yeh W-H, Pan B, Hu Y-J, Hu JH, Thompson DB, Shu Y, Li Y, Wang H, Yang S, Xu Q, Polley DB, Liberman MC, Kong W-J, Holt JR, Chen Z-Y, Liu DR, Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents, Nature 553 (2018) 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Liu Q, Wang X, Liu X, Kumar S, Gochman G, Ji Y, Liao Y-P, Chang CH, Situ W, Lu J, Jiang J, Mei K-C, Meng H, Xia T, Nel AE, Use of polymeric nanoparticle platform targeting the liver to induce Treg-mediated antigen-specific immune tolerance in a pulmonary allergen sensitization model, ACS Nano 13 (2019) 4778–4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Liu Q, Wang X, Liu X, Liao Y-P, Chang CH, Mei K-C, Jiang J, Tseng S, Gochman G, Huang M, Thatcher Z, Li J, Allen SD, Lucido L, Xia T, Nel AE, Antigen- and epitope-delivering nanoparticles targeting liver induce comparable Immunotolerance in allergic airway disease and anaphylaxis as nanoparticle-delivering pharmaceuticals, ACS Nano 15 (2021) 1608–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Cifuentes-Rius A, Desai A, Yuen D, Johnston APR, Voelcker NH, Inducing immune tolerance with dendritic cell-targeting nanomedicines, Nat. Nanotechnol 16 (2021) 37–46. [DOI] [PubMed] [Google Scholar]
- [195].Pattipeiluhu R, Arias-Alpizar G, Basha G, Chan KYT, Bussmann J, Sharp TH, Moradi MA, Sommerdijk N, Harris EN, Cullis PR, Kros A, Witzigmann D, Campbell F, Anionic lipid nanoparticles preferentially deliver mRNA to the hepatic reticuloendothelial system, Adv. Mater 34 (2022) e2201095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Veiga N, Goldsmith M, Granot Y, Rosenblum D, Dammes N, Kedmi R, Ramishetti S, Peer D, Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes, Nat. Commun 9 (2018) 4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Rurik JG, Tombácz I, Yadegari A, Méndez Fernández PO, Shewale SV, Li L, Kimura T, Soliman OY, Papp TE, Tam YK, Mui BL, Albelda SM, Puré E, June CH, Aghajanian H, Weissman D, Parhiz H, Epstein JA, CAR T cells produced in vivo to treat cardiac injury, Science 375 (2022) 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Robinson EL, Port JD, Utilization and potential of RNA-based therapies in cardiovascular disease, JACC: Basic to Translational Science 7 (2022) 956–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Hebert PD, Cywinska A, Ball SL, deWaard JR, Biological identifications through DNA barcodes, Proc. Biol. Sci 270 (2003) 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Guimaraes PPG, Zhang R, Spektor R, Tan M, Chung A, Billingsley MM, El-Mayta R, Riley RS, Wang L, Wilson JM, Mitchell MJ, Ionizable lipid nanoparticles encapsulating barcoded mRNA for accelerated in vivo delivery screening, J. Control. Release 316 (2019) 404–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Willmann JK, van Bruggen N, Dinkelborg LM, Gambhir SS, Molecular imaging in drug development, Nat. Rev. Drug Discov 7 (2008) 591–607. [DOI] [PubMed] [Google Scholar]
- [202].Burdette D, Ci L, Shilliday B, Slauter R, Auerbach A, Kenney M, Almarsson Ö, Cheung E, Hendrick T, Systemic exposure, metabolism, and elimination of [14C]-labeled amino lipid, lipid 5, after a single administration of mRNA encapsulating lipid nanoparticles to Sprague-Dawley rats, Drug Metab. Dispos 51 (2023) 804–812. [DOI] [PubMed] [Google Scholar]
- [203].Cheung BCL, Sun THT, Leenhouts JM, Cullis PR, Loading of Doxorubicin into Liposomes by Forming Mn2+ − Drug Complexes, Biochimica et Biophysica Acta (BBA) - Biomembranes 1414 (1998) 205–216. [DOI] [PubMed] [Google Scholar]
- [204].Mei K-C, Thota N, Wei P-S, Yi B, Bonacquisti EE, Nguyen J, Calreticulin P-domain-derived “eat-me” peptides for enhancing liposomal uptake in dendritic cells, Int. J. Pharm 653 (2024) 123844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H, A robust and high-throughput Cre reporting and characterization system for the whole mouse brain, Nat. Neurosci 13 (2010) 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Lokugamage MP, Sago CD, Dahlman JE, Testing thousands of nanoparticles in vivo using DNA barcodes, current opinion in biomedical, Engineering 7 (2018) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Dahlman JE, Kauffman KJ, Xing Y, Shaw TE, Mir FF, Dlott CC, Langer R, Anderson DG, Wang ET, Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics, Proc. Natl. Acad. Sci 114 (2017) 2060–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Huayamares SG, Lokugamage MP, Rab R, Da Silva Sanchez AJ, Kim H, Radmand A, Loughrey D, Lian L, Hou Y, Achyut BR, Ehrhardt A, Hong JS, Sago CD, Paunovska K, Echeverri ES, Vanover D, Santangelo PJ, Sorscher EJ, Dahlman JE, High-throughput screens identify a lipid nanoparticle that preferentially delivers mRNA to human tumors in vivo, J. Control. Release 357 (2023) 394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Da Silva Sanchez AJ, Zhao K, Huayamares SG, Hatit MZC, Lokugamage MP, Loughrey D, Dobrowolski C, Wang S, Kim H, Paunovska K, Kuzminich Y, Dahlman JE, Substituting racemic ionizable lipids with stereopure ionizable lipids can increase mRNA delivery, J. Control. Release 353 (2023) 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Lokugamage MP, Gan Z, Zurla C, Levin J, Islam FZ, Kalathoor S, Sato M, Sago CD, Santangelo PJ, Dahlman JE, Mild innate immune activation overrides efficient nanoparticle-mediated RNA delivery, Adv. Mater 32 (2020) 1904905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Rhym LH, Manan RS, Koller A, Stephanie G, Anderson DG, Peptide-encoding mRNA barcodes for the high-throughput in vivo screening of libraries of lipid nanoparticles for mRNA delivery, Nature Biomed. Eng 7 (2023) 901–910. [DOI] [PubMed] [Google Scholar]
- [212].Brenner S, Jacob F, Meselson M, An unstable intermediate carrying information from genes to ribosomes for protein synthesis, Nature 190 (1961) 576–581. [DOI] [PubMed] [Google Scholar]
- [213].Gros F, Hiatt H, Gilbert W, Kurland CG, Risebrough RW, Watson JD, Unstable ribonucleic acid revealed by pulse labelling of Escherichia Coli, Nature 190 (1961) 581–585. [DOI] [PubMed] [Google Scholar]
- [214].Jacob F, Monod J, Genetic regulatory mechanisms in the synthesis of proteins, J. Mol. Biol 3 (1961) 318–356. [DOI] [PubMed] [Google Scholar]
- [215].Lockard RE, Lingrel JB, The synthesis of mouse hemoglobin chains in a rabbit reticulocyte cell-free system programmed with mouse reticulocyte 9S RNA, Biochem. Biophys. Res. Commun 37 (1969) 204–212. [DOI] [PubMed] [Google Scholar]
- [216].Lim L, Canellakis ES, Adenine-rich polymer associated with rabbit reticulocyte messenger RNA, Nature 227 (1970) 710–712. [DOI] [PubMed] [Google Scholar]
- [217].Furuichi Y, Miura K-I, A blocked structure at the 5′ terminus of mRNA from cytoplasmic polyhedrosis virus, Nature 253 (1975) 374–375. [DOI] [PubMed] [Google Scholar]
- [218].Melton DA, Krieg PA, Rebagliati MR, Maniatis T, Zinn K, Green MR, Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter, Nucleic Acids Res. 12 (1984) 7035–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL, Direct gene transfer into mouse muscle in vivo, Science 247 (1990) 1465–1468. [DOI] [PubMed] [Google Scholar]
- [220].Jirikowski GF, Sanna PP, Maciejewski-Lenoir D, Bloom FE, Reversal of diabetes insipidus in Brattleboro rats: intrahypothalamic injection of vasopressin mRNA, Science 255 (1992) 996–998. [DOI] [PubMed] [Google Scholar]
- [221].Martinon F, Krishnan S, Lenzen G, Magné R, Gomard E, Guillet JG, Lévy JP, Meulien P, Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA, Eur. J. Immunol 23 (1993) 1719–1722. [DOI] [PubMed] [Google Scholar]
- [222].Conry RM, LoBuglio AF, Wright M, Sumerel L, Pike MJ, Johanning F, Benjamin R, Lu D, Curiel DT, Characterization of a messenger RNA polynucleotide vaccine vector, Cancer Res. 55 (1995) 1397–1400. [PubMed] [Google Scholar]
- [223].Boczkowski D, Nair SK, Snyder D, Gilboa E, Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo, J. Exp. Med 184 (1996) 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Karikó K,Kuo A, Barnathan ES, Overexpression of urokinase receptor in mammalian cells following administration of the in vitro transcribed encoding mRNA, Gene Ther. 6 (1999) 1092–1100. [DOI] [PubMed] [Google Scholar]
- [225].Zhou WZ, Hoon DS, Huang SK, Fujii S, Hashimoto K, Morishita R, Kaneda Y, RNA melanoma vaccine: induction of antitumor immunity by human glycoprotein 100 mRNA immunization, Hum. Gene Ther 10 (1999) 2719–2724. [DOI] [PubMed] [Google Scholar]
- [226].Karikó K, Buckstein M, Ni H, Weissman D, Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA, Immunity 23 (2005) 165–175. [DOI] [PubMed] [Google Scholar]
- [227].Krammer F, Palese P, Profile of Katalin Karikó and drew Weissman: 2023 Nobel laureates in physiology or medicine, Proc. Natl. Acad. Sci 121 (2024) e2400423121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Hamilton AJ, Baulcombe DC, A species of small antisense RNA in posttranscriptional gene silencing in plants, Science 286 (1999) 950–952. [DOI] [PubMed] [Google Scholar]
- [229].Ahn I, Kang CS, Han J, Where should siRNAs go: applicable organs for siRNA drugs, Exp. Mol. Med 55 (2023) 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, McDermott A, Flach B, Doria-Rose NA, Corbett KS, Morabito KM, O’Dell S, Schmidt SD, Swanson PA 2nd, Padilla M, Mascola JR, Neuzil KM, Bennett H, Sun W, Peters E, Makowski M, Albert J, Cross K, Buchanan W, Pikaart-Tautges R, Ledgerwood JE, Graham BS, Beigel JH, An mRNA vaccine against SARS-CoV-2 - preliminary report, N. Engl. J. Med 383 (2020) 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Ye Z, Harmon J, Ni W, Li Y, Wich D, Xu Q, The mRNA vaccine revolution: COVID-19 has launched the future of vaccinology, ACS Nano 17 (2023) 15231–15253. [DOI] [PubMed] [Google Scholar]
- [232].Feldman RA, Fuhr R, Smolenov I, Mick Ribeiro A, Panther L, Watson M, Senn JJ, Smith M, Almarsson Ӧ, Pujar HS, Laska ME, Thompson J, Zaks T, Ciaramella G, mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials, Vaccine 37 (2019) 3326–3334. [DOI] [PubMed] [Google Scholar]
- [233].Fortner A, Bucur O, mRNA-based vaccine technology for HIV, Discoveries (Craiova) 10 (2022) e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Shaw CA, August A, Bart S, Booth PJ, Knightly C, Brasel T, Weaver SC, Zhou H, Panther L, A phase 1, randomized, placebo-controlled, dose-ranging study to evaluate the safety and immunogenicity of an mRNA-based chikungunya virus vaccine in healthy adults, Vaccine 41 (2023) 3898–3906. [DOI] [PubMed] [Google Scholar]
- [235].Chen GL, Mithani R, Kapoor A, Lu S, Asmar LE, Panozzo CA, Shaw CA, Stoszek SK, August A, 234. Safety and immunogenicity of mRNA-1345, an mRNA-based RSV vaccine in younger and older adult cohorts: results from a phase 1, randomized clinical trial, open forum, Infect. Dis 9 (Suppl. 2) (2022) ofac492.312. [Google Scholar]
- [236].Aldrich C, Leroux-Roels I, Huang KB, Bica MA, Loeliger E, Schoenborn-Kellenberger O, Walz L, Leroux-Roels G, von Sonnenburg F, Oostvogels L, Proof-of-concept of a low-dose unmodified mRNA-based rabies vaccine formulated with lipid nanoparticles in human volunteers: a phase 1 trial, Vaccine 39 (2021) 1310–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Zhao G, Ho W, Chu J, Xiong X, Hu B, Boakye-Yiadom KO, Xu X, Zhang X-Q, Inhalable siRNA nanoparticles for enhanced tumor-targeting treatment of KRAS-mutant non-small-cell lung Cancer, ACS Appl. Mater. Interfaces 15 (2023) 31273–31284. [DOI] [PubMed] [Google Scholar]
- [238].Taniuchi K, Yawata T, Tsuboi M, Ueba T, Saibara T, Efficient delivery of small interfering RNAs targeting particular mRNAs into pancreatic cancer cells inhibits invasiveness and metastasis of pancreatic tumors, Oncotarget 10 (2019) 2869–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Gu S, Ngamcherdtrakul W, Reda M, Hu Z, Gray JW, Yantasee W, Lack of acquired resistance in HER2-positive breast cancer cells after long-term HER2 siRNA nanoparticle treatment, PloS One 13 (2018) e0198141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Oner E, Kotmakci M, Baird A-M, Gray SG, Debelec Butuner B, Bozkurt E, Kantarci AG, Finn SP, Development of EphA2 siRNA-loaded lipid nanoparticles and combination with a small-molecule histone demethylase inhibitor in prostate cancer cells and tumor spheroids, J. Nanobiotechnol 19 (2021) 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Singh MS, Ramishetti S, Landesman-Milo D, Goldsmith M, Chatterjee S, Palakuri R, Peer D, Therapeutic gene silencing using targeted lipid nanoparticles in metastatic ovarian Cancer, Small 17 (2021) 2100287. [DOI] [PubMed] [Google Scholar]
- [242].Tabernero J, Shapiro GI, LoRusso PM, Cervantes A, Schwartz GK, Weiss GJ, Paz-Ares L, Cho DC, Infante JR, Alsina M, Gounder MM, Falzone R, Harrop J, White AC, Toudjarska I, Bumcrot D, Meyers RE, Hinkle G, Svrzikapa N, Hutabarat RM, Clausen VA, Cehelsky J, Nochur SV, Gamba-Vitalo C, Vaishnaw AK, Sah DW, Gollob JA, Burris HA 3rd, First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement, Cancer Discov. 3 (2013) 406–417. [DOI] [PubMed] [Google Scholar]
- [243].Jasiewicz NE, Mei K-C, Oh HM, Chansoria P, Hendy DA, Bonacquisti EE, Bachelder EM, Ainslie KM, Yin H, Qian L, Jensen BC, Nguyen J, ZipperCells exhibit enhanced accumulation and retention at the site of myocardial infarction, Adv. Healthc. Mater 12 (2023) 2201094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Jackson AL, Linsley PS, Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application, Nat. Rev. Drug Discov 9 (2010) 57–67. [DOI] [PubMed] [Google Scholar]
- [245].Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC, Hughes CM, Shanmugam KS, Bhattacharjee A, Meyerson M, Collins FS, Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells, Proc. Natl. Acad. Sci. U. S. A 101 (2004) 1892–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Yang T, Li C, Wang X, Zhao D, Zhang M, Cao H, Liang Z, Xiao H, Liang XJ, Weng Y, Huang Y, Efficient hepatic delivery and protein expression enabled by optimized mRNA and ionizable lipid nanoparticle, Bioact Mater 5 (2020) 1053–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Bicknell AA, Reid DW, Licata MC, Jones AK, Cheng YM, Li M, Hsiao CJ, Pepin CS, Metkar M, Levdansky Y, Fritz BR, Andrianova EA, Jain R, Valkov E, Köhrer C, Moore MJ, Attenuating ribosome load improves protein output from mRNA by limiting translation-dependent mRNA decay, Cell Rep. 43 (2024) 114098. [DOI] [PubMed] [Google Scholar]
- [248].Boo SH, Kim YK, The emerging role of RNA modifications in the regulation of mRNA stability, Exp. Mol. Med 52 (2020) 400–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Raabe V, Bailey R, Swanson KA, Li P, Koury K, Kalina W, Cooper D, Fontes-Garfias C, Shi P-Y, Türeci Ö, Tompkins KR, Walsh EE, Frenck R, Falsey AR, Dormitzer PR, Gruber WC, Șahin U, Jansen KU, Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults, Nature 586 (2020) 589–593. [DOI] [PubMed] [Google Scholar]
- [250].Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, Himansu S, Schäfer A, Ziwawo CT, DiPiazza AT, Dinnon KH, Elbashir SM, Shaw CA, Woods A, Fritch EJ, Martinez DR, Bock KW, Minai M, Nagata BM, Hutchinson GB, Wu K, Henry C, Bahl K, Garcia-Dominguez D, Ma L, Renzi I, Kong W-P, Schmidt SD, Wang L, Zhang Y, Phung E, Chang LA, Loomis RJ, Altaras NE, Narayanan E, Metkar M, Presnyak V, Liu C, Louder MK, Shi W, Leung K, Yang ES, West A, Gully KL, Stevens LJ, Wang N, Wrapp D, Doria-Rose NA, Stewart-Jones G, Bennett H, Alvarado GS, Nason MC, Ruckwardt TJ, McLellan JS, Denison MR, Chappell JD, Moore IN, Morabito KM, Mascola JR, Baric RS, Carfi A, Graham BS, SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness, Nature 586 (2020) 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Kenjo E, Hozumi H, Makita Y, Iwabuchi KA, Fujimoto N, Matsumoto S, Kimura M, Amano Y, Ifuku M, Naoe Y, Inukai N, Hotta A, Low immunogenicity of LNP allows repeated administrations of CRISPR-Cas9 mRNA into skeletal muscle in mice, Nat. Commun 12 (2021) 7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Miao L, Li L, Huang Y, Delcassian D, Chahal J, Han J, Shi Y, Sadtler K, Gao W, Lin J, Doloff JC, Langer R, Anderson DG, Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation, Nat. Biotechnol 37 (2019) 1174–1185. [DOI] [PubMed] [Google Scholar]
- [253].Mei K-C, Stiepel RT, Bonacquisti E, Jasiewicz NE, Chaudhari AP, Tiwade PB, Bachelder EM, Ainslie KM, Fenton OS, Nguyen J, Single-tailed heterocyclic carboxamide lipids for macrophage immune-modulation, biomaterials, Science 11 (2023) 2693–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Chen S, Zaifman J, Kulkarni JA, Zhigaltsev IV, Tam YK, Ciufolini MA, Tam YYC, Cullis PR, Dexamethasone prodrugs as potent suppressors of the immunostimulatory effects of lipid nanoparticle formulations of nucleic acids, J. Control. Release 286 (2018) 46–54. [DOI] [PubMed] [Google Scholar]
- [255].Omo-Lamai S, Wang Y, Patel MN, Essien E-O, Shen M, Majumdar A, Espy C, Wu J, Channer B, Tobin M, Murali S, Papp TE, Maheshwari R, Wang L, Chase LS, Zamora ME, Arral ML, Marcos-Contreras OA, Myerson JW, Hunter CA, Tsourkas A, Muzykantov V, Brodsky I, Shin S, Whitehead KA, Gaskill P, Discher D, Parhiz H, Brenner JS, Lipid Nanoparticle-Associated Inflammation is Triggered by Sensing of Endosomal Damage: Engineering Endosomal Escape Without Side Effects, bioRxiv (2024), 2024.2004.2016.589801. [Google Scholar]
- [256].Naddaf M, First trial of ‘base editing’ in humans lowers cholesterol — but raises safety concerns, Nature 623 (2024) 671–672. [DOI] [PubMed] [Google Scholar]
- [257].Kim J, Jozic A, Mukherjee A, Nelson D, Chiem K, Khan MSR, Torrelles JB, Martinez-Sobrido L, Sahay G, Rapid generation of circulating and mucosal decoy human ACE2 using mRNA Nanotherapeutics for the potential treatment of SARS-CoV-2, Adv. Sci 9 (2022) 2202556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Wang Y, Tiruthani K, Li S, Hu M, Zhong G, Tang Y, Roy S, Zhang L, Tan J, Liao C, Liu R, mRNA delivery of a bispecific single-domain antibody to polarize tumor-associated macrophages and synergize immunotherapy against liver malignancies, Adv. Mater 33 (2021) e2007603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Kong N, Tao W, Ling X, Wang J, Xiao Y, Shi S, Ji X, Shajii A, Gan ST, Kim NY, Duda DG, Xie T, Farokhzad OC, Shi J, Synthetic mRNA nanoparticle-mediated restoration of p53 tumor suppressor sensitizes p53-deficient cancers to mTOR inhibition, Sci. Transl. Med 11 (2019) eaaw1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Sago CD, Lokugamage MP, Paunovska K, Vanover DA, Monaco CM, Shah NN, Gamboa Castro M, Anderson SE, Rudoltz TG, Lando GN, Munnilal Tiwari P, Kirschman JL, Willett N, Jang YC, Santangelo PJ, Bryksin AV, Dahlman JE, High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing, Proc. Natl. Acad. Sci 115 (2018) E9944–E9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Paunovska K, Sago CD, Monaco CM, Hudson WH, Castro MG, Rudoltz TG, Kalathoor S, Vanover DA, Santangelo PJ, Ahmed R, Bryksin AV, Dahlman JE, A direct comparison of in vitro and in vivo nucleic acid delivery mediated by hundreds of nanoparticles reveals a weak correlation, Nano Lett. 18 (2018) 2148–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Paunovska K, Gil CJ, Lokugamage MP, Sago CD, Sato M, Lando GN, Gamboa Castro M, Bryksin AV, Dahlman JE, Analyzing 2000 In vivo drug delivery data points reveals cholesterol structure impacts nanoparticle delivery, ACS Nano 12 (2018) 8341–8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Guimarães PPG, Figueroa-Espada CG, Riley RS, Gong N, Xue L, Sewastianik T, Dennis PS, Loebel C, Chung A, Shepherd SJ, Haley RM, Hamilton AG, El-Mayta R, Wang K, Langer R, Anderson DG, Carrasco RD, Mitchell MJ, In vivo bone marrow microenvironment siRNA delivery using lipid–polymer nanoparticles for multiple myeloma therapy, Proc. Natl. Acad. Sci 120 (2023) e2215711120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Da Silva Sanchez AJ, Dobrowolski C, Cristian A, Echeverri ES, Zhao K, Hatit MZC, Loughrey D, Paunovska K, Dahlman JE, Universal barcoding predicts in vivo ApoE-independent lipid nanoparticle delivery, Nano Lett. 22 (2022) 4822–4830. [DOI] [PubMed] [Google Scholar]
- [265].Radmand A, Lokugamage MP, Kim H, Dobrowolski C, Zenhausern R, Loughrey D, Huayamares SG, Hatit MZC, Ni H, Del Cid A, Da Silva Sanchez AJ, Paunovska K, Schrader Echeverri E, Shajii A, Peck H, Santangelo PJ, Dahlman JE, The transcriptional response to lung-targeting lipid nanoparticles in vivo, Nano Lett. 23 (2023) 993–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Paunovska K, Da Silva Sanchez AJ, Sago CD, Gan Z, Lokugamage MP, Islam FZ, Kalathoor S, Krupczak BR, Dahlman JE, Nanoparticles Containing Oxidized Cholesterol Deliver mRNA to the Liver Microenvironment at Clinically Relevant Doses, Advanced materials (Deerfield Beach, Fla.) 31 (2019) e1807748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Guimaraes PPG, Zhang R, Spektor R, Tan M, Chung A, Billingsley MM, El-Mayta R, Riley RS, Wang L, Wilson JM, Mitchell MJ, Ionizable lipid nanoparticles encapsulating barcoded mRNA for accelerated in vivo delivery screening, J. Control. Release 316 (2019) 404–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Hardianto A, Muscifa ZS, Widayat W, Yusuf M, Subroto T, The effect of ethanol on lipid nanoparticle stabilization from a molecular dynamics simulation perspective, Molecules 28 (2023) 4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Jasiewicz NE, C Mei K, Oh HM, et al. , In situ-crosslinked Zippersomes enhance cardiac repair by increasing accumulation and retention, Bioeng Transl Med (2024) e10697.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Li Yue-ting, Mei Kuo-Ching, Revadee Liam-Or Julie Tzu-Wen Wang, Faruqu Farid N., Zhu Shengzhang, Wang Yong-lin, Lu Yuan, Al-Jamal Khuloud T., Graphene Oxide Nanosheets Toxicity in Mice Is Dependent on Protein Corona Composition and Host Immunity, ACS Nano 18 (2024) 22572–22585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.


