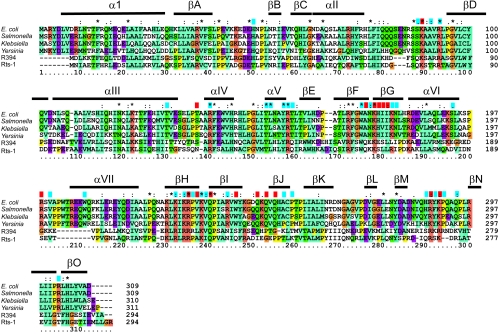
FIG. 12.
Sequence alignment of some Tus and Tus-like proteins. An alignment of the Tus protein sequences from E. coli, Salmonella enterica serovar Typhimurium, Klebsiella pneumoniae subsp. ozaenae, and Yersinia pestis, along with sequences of Tus-like proteins from the R394 plasmid of S. enterica serovar Typhimurium and the Rts-1 plasmid of Proteus vulgaris, was carried out and colored using the default parameters in CLUSTAL_X (64). Essentially, residues are colored where more than a given percentage of residues belong to one class: cyan, aliphatic and hydrophobic residues; orange, basic residues; purple, acidic residues; green, neutral hydrogen bonding residues. All glycines are colored brown, and all prolines are colored yellow. Secondary structure elements from the Tus-Ter crystal structure are shown above the alignment. Residues that make DNA backbone contacts in the crystal structure are shown with a blue block above the alignment. Those residues that make sequence-specific contacts with the Ter DNA are shown with a red block. Tus and Tus-like proteins were identified using PSI-BLAST (4).