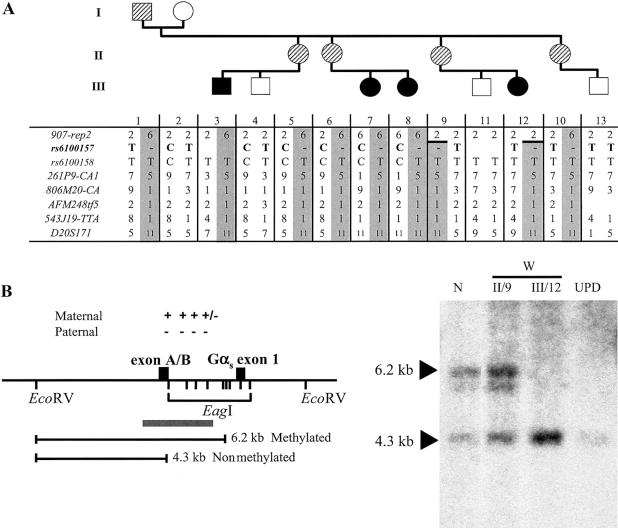
Figure 2.
A, Evidence of allelic loss within the STX16 locus in the obligate carriers W-II/5 and W-II/6. All the affected individuals (blackened symbols) as well as the obligate carriers (striped symbols) share the same disease-associated haplotype (gray columns) at 20q13.3; note that not all investigated markers are shown. A recombination event occurred in individual W-II/9 between markers 907-rep2 and 261P9-CA1 and was transmitted to her affected daughter, W-III/12, thus reducing the critical interval likely containing the genetic lesion that causes AD-PHP-Ib. Evidence of allelic loss was found at SNP rs6100157 (bold typeface) in obligate carriers W-II/5 and W-II/6. B, Loss of methylation at GNAS exon A/B in the newborn W-III/12. The methylation status of GNAS exon A/B was assessed by Southern blot analysis of cord blood DNA from patient W-III/12 and of genomic DNA from her unaffected mother (W-II/9), an individual with paternal uniparental isodisomy of chromosome 20q (“UPD”) (Bastepe et al. 2001a), and a healthy control individual (“N”). These samples were digested with a methylation-sensitive (EagI) and a methylation-insensitive (EcoRV) restriction endonuclease, as described elsewhere (Bastepe et al. 2001b). Locations of restriction sites and their methylation status (“+” = methylated; “−” = nonmethylated), the predicted fragments, and the probe (gray bar) are shown in the left panel. Affected individuals (UPD and W-III/12) show loss of methylation at exon A/B, whereas unaffected individuals (N and W-II/9) have one methylated and one nonmethylated allele. Fragment sizes are indicated by arrowheads. Note that the faint hybridizing band below the 6.2-kb band reflects partial methylation (“+/−”) and, hence, incomplete digestion at one of the EagI sites between exon A/B and Gαs exon 1.