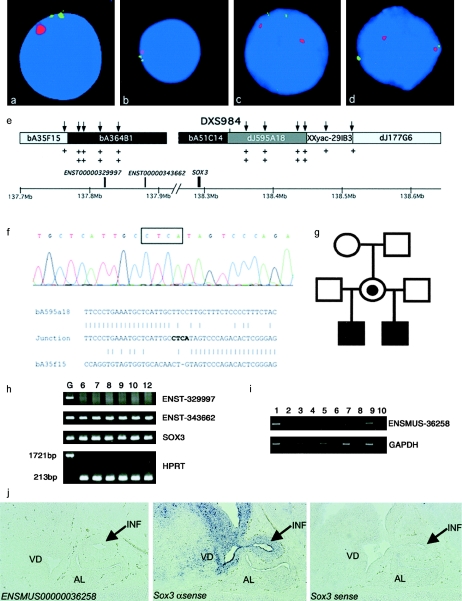
Figure 2.
Duplication of SOX3 in family A. a–d, Representative interphase nuclei from four members of the family (older sibling [a], younger half brother [b], mother [c], and maternal grandmother [d]), with bA51C14 shown in green and the X centromere shown in red. e, Data used to map the extent of the duplication. Clones from the SOX3 genomic region are shown along with position (in Mb) on the X chromosome (NCBI build 34 assembly of the human genome). The clones duplicated by interphase FISH are blackened, those not duplicated are shaded light gray, and the clone containing the breakpoint is shaded dark gray; unknown duplication status is indicated by the unblackened area. The positions of genes within the duplicated region are designated by blackened boxes above the scale bar. The positions of UPQFM-PCR primer pairs are designated by vertical arrows above the contig. Two plus signs underneath a primer pair indicate that dosage quotients for this primer pair were consistent with a duplication; one plus sign shows that this region had a normal copy number. The position of the informative polymorphic marker, DXS984, is also shown. f, Electropherogram showing sequence from the duplication breakpoint in family A. The four nucleotides not present in either the proximal or the distal normal sequence are boxed above the electropherogram. Underneath, a ClustalW alignment of the normal distal (bA595A18), normal proximal (bA35F15), and junction sequence is shown, demonstrating that there is no homology at the proximal and distal ends of the duplication. g, Pedigree of family A. h, RT-PCR for the three genes within the duplication, performed using human brain mRNA at 6, 7, 8, 9, 10, and 12 wk of gestation. Genomic DNA was included as a positive control. Transcripts were present for SOX3 and a novel gene (Ensembl ENST00000343662). The larger 1,721-bp product was not amplified from the brain RNA, showing that there was no genomic DNA contamination. i, RT-PCR analysis for ENSMUSG00000036258 (murine homologue of ENSG00000343662), showing expression in the mouse embryo at 14.5 days postcoitum (dpc). Expression was not detected in the hypothalamic/pituitary region, the pituitary progenitor cell line aT1-1, or the adult pituitary. No amplification was evident in the −RT controls, indicating that the cDNA was not contaminated with genomic DNA. Lane 1, 14.5–dpc embryo +RT; lane 2, 14.5-dpc embryo −RT; lane 3, 14.5-dpc hypothalamus/pituitary +RT; lane 4, 14.5-dpc hypothalamus/pituitary −RT; lane 5, aT1-1 +RT; lane 6, aT1-1 −RT; lane 7, adult pituitary +RT; lane 8, adult pituitary −RT; lane 9, genomic DNA as template; lane 10, water as a blank. j, In situ analysis of the ENSMUSG00000036258 and Sox3 genes in the developing murine hypothalamus, infundibulum, and pituitary. No expression of ENSMUSG00000036258 was detected (left panel). Sox3 is expressed in the infundibulum (arrow) and ventral diencephalon but not in the presumptive anterior pituitary (middle panel). No staining was observed using the Sox3 sense control probe (right panel). INF = infundibulum; VD = ventral diencephalon; AL = anterior lobe.