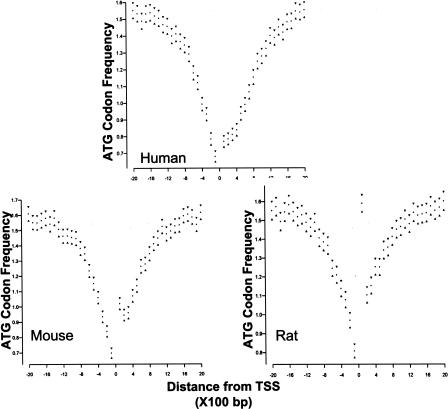
Figure 4.
A subclass of promoters with ATG deserts is identified in human, mouse, and rat genomes by a computational algorithm. Analysis of 8003 genes that could be aligned among the human, rat, and mouse genomes identified 6595 genes as having potential ATG desert. Validation of the prediction was by plotting ATG frequencies within 2 kb of the major TSS, for each of the species. The major TSS for any given gene is located at position 0 on the X axis. Each point represents the frequency of observed ATG codons within a 100-bp window; 95% confidence intervals are indicated by the triangles. (The high ATG count in window 1 in rat and moderately high ATG count in mouse are the result of genome annotations, which defined the first ATG of an open reading frame as the transcription start site for many of the rat genes and some of the mouse genes.)