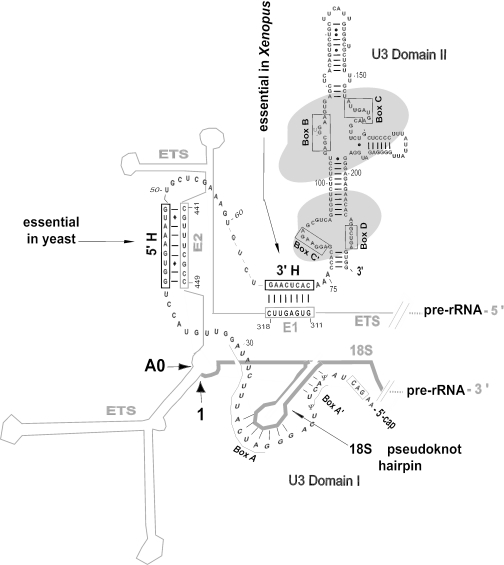
Figure 2.
U3 snoRNA interaction with the ETS. Base pairing between the 3′-hinge (3′H) of U3 snoRNA and region E1 of the pre-rRNA ETS is critical and sufficient for rRNA processing in Xenopus, whereas base pairing between the U3 5′-hinge (5′H) and region E2 of the ETS is auxiliary (26). In contrast, the 5′H–E2 interaction is essential in yeast and the 3′H–E1 interaction is not sufficient to support rRNA processing (27). Domain II of U3 snoRNA has many base-paired stems and binds several proteins (indicated by shading). Domain I of U3 is shown in an open configuration that can interact with the 5′ end of the 18S region of pre-rRNA (9,26,28). The conserved sequences comprising boxes that are conserved in U3 snoRNA from all species are indicated. Base pairing of U3 box A with the first terminal loop in 18S rRNA is thought to prevent premature pseudoknot formation (29,30). The pre-rRNA cleavage sites A0 and 1 are close to one another in the proposed secondary structure of the ETS (9,31).