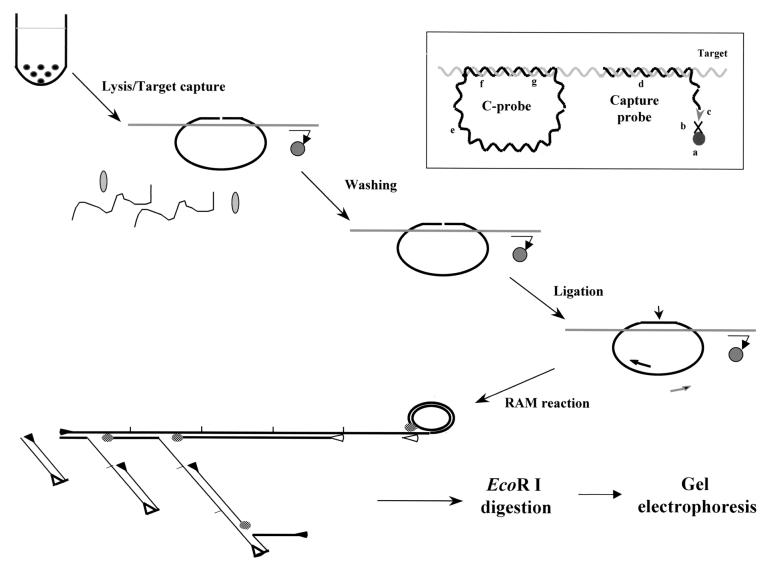
FIG. 1.
Schematic representation of the RAM assay. Target DNA, capture probe, C-probe, and paramagnetic bead are added to hybridization buffer to allow formation of a hybrid complex. The hybrid is captured on a paramagnetic bead, allowing extensive washing to remove unbound C-probe and cellular components. The C-probe aligned on the target is linked together by a DNA ligase. RAM amplification is then carried out by the addition of forward (▸) and reverse (◃) primers and DNA polymerase (shaded ovals). The forward primer bound to the C-probe is extended by the polymerase and continues after one round of synthesis by displacing the bound forward primer and its extended product, generating a long ssDNA with repeated sequence. With the reactions going on, multiple reverse primers can bind to the nascent ssDNA as their binding sites are available. Each bound reverse primer will be extended and displace the downstream primers and their extended products. The forward primer binding sites of the displaced ssDNA are then available for the forward primer to bind and extend similarly, thus forming a large ramifying DNA complex. Finally, the RAM products are examined by gel electrophoresis after EcoRI digestion. Insert: the C-probe hybridizes to the target through their complementary regions (f and g), and sequence at the noncomplementary region (e) is generic for the binding of primers. The C-probe-target hybrid is captured on a paramagnetic bead (a) through the binding of the biotin moiety (c) of the capture probe (d) to streptavidin (b) coated on the beads.