Figure 1.
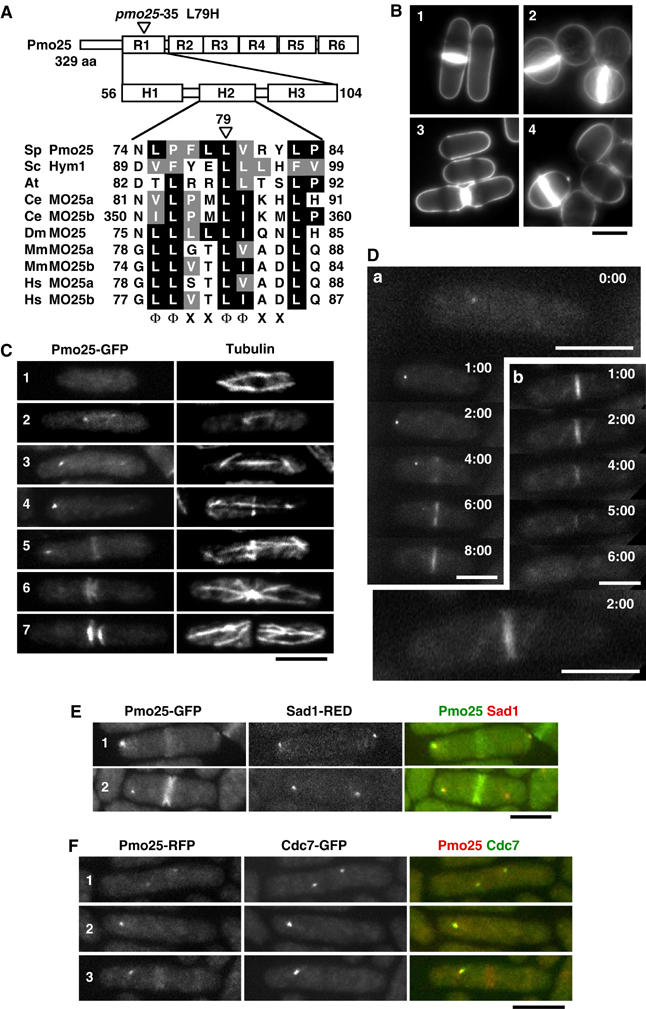
Structure and localization of Pmo25. (A) Pmo25 consists of six repeats (R1–R6) each having three helices (H1–H3). The mutation site of the pmo25-35 allele is located within H2 of R1. The H2 of Pmo25 (NCBI accession no. NP_594685) is aligned with Saccharomyces cerevisiae Sc Hym1 (NP_012732), Arabidopsis thaliana (CAB78730), C. elegans Ce MO25a (CAB16486), Ce MO25b (NP_508691), Drosophila melanogaster Dm MO25 (P91891), Mus musculus Mm MO25a (NP_598542), Mm MO25b (AAH16128), Homo sapiens Hs MO25a (NP_057373), and Hs MO25b (CAC37735). The conserved motif (Φ, hydrophobic; X, any residue) is shown. (B) Calcofluor staining of WT cells (1), pmo25-deleted cells (2), pmo25-deleted cells with pREP41-Pmo25 ((3) in EMM, induced condition; (4) in EMM containing thiamine, repressed condition). (C, D) WT cells having pmo25+:GFP:kanr grown in YES medium at 25°C were stained with anti-tubulin antibody (C). Pmo25 localization at mitosis (a) and cell separation (b) of the living cells was observed by time lapse (minutes) (D). (E, F) WT cells having pmo25+:GFP:kanr and sad1+:dsRED:LEU2 (E) or pmo25+:mRFP:kanr and cdc7+:GFP:kanr (F) grown in YES medium at 25°C were used. Bars indicate 5 μm.
