Fig. 2.
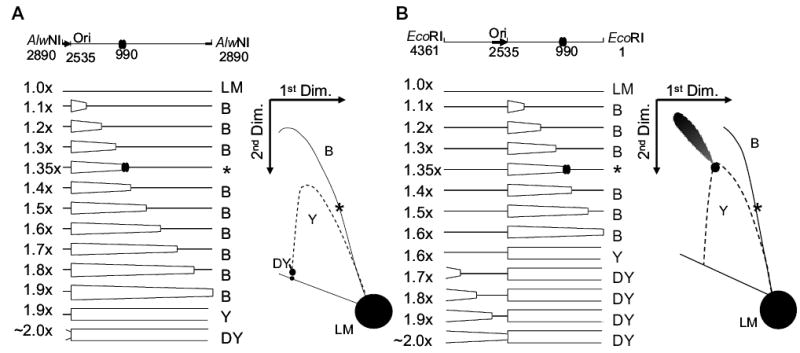
Diagrams of pBR322 DNA after cleavage with AlwNI (A) or EcoRI (B). Replication of pBR322 begins unidirectionally at position 2535 and is indicated by the thick black arrow. AlwNI cuts just behind the origin at position 2890 and EcoRI cuts midway around the plasmid at position 1, as indicated in the schematic on top. The major gyrase binding site is at position 990 and is indicated by two overlapping ovals. The map indicates the forms of DNA produced as replication proceeds from 1.0× to 2.0×. Non-replicating linear (LM) monomers are 4361 base pairs. As the replication fork crosses the location of the restriction endonuclease recognition site, the resulting molecules change from bubble-form (B) to double-Y form (DY), by passing through a simple-Y form [expected as a spot along the simple-Y arc (Y)]. The bubble-to-double-Y transition occurs at 1.9× for AlwNI and 1.6× for EcoRI. At 1.35×, the replication fork reaches the gyrase binding site centred at 990 bp, where stalling can occur if a gyrase cleavage complex is bound (molecules indicated by an asterisk). To the right of the maps are illustrations of the expected 2D gel electrophoresis patterns. The location of the entire simple-Y arc is shown as a dotted line; this arc is not expected for normal pBR322 replication (except for the discrete spot at the bubble-to-double-Y transition).
