Fig. 3.
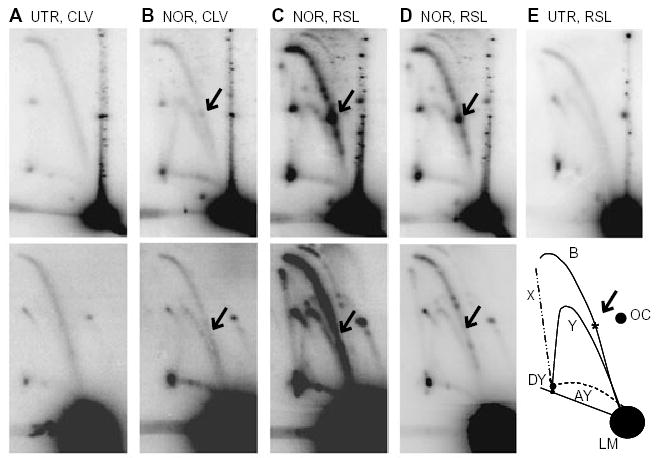
Norfloxacin treatment causes stalled replication forks at gyrase binding sites. DNA was isolated from JH39 cells carrying pBR322 (upper panels) or pBR322MUT990 (lower panels) and digested with AlwNI. The samples were as follows:
A. Cells were untreated (UTR) and DNA was isolated under cleavage conditions (CLV).
B. Cells were treated with norfloxacin (NOR) and DNA was isolated under cleavage conditions.
C. Cells were treated with norfloxacin and DNA was isolated under resealing conditions (RSL).
D. Shorter exposures of panel C.
E. Cells were untreated and DNA was isolated under resealing conditions (upper panel). The illustration (E, lower) indicates the various arcs seen after AlwNI digestion where B, bubble; LM, linear monomer; Y, simple-Y; AY, asymmetric-Y; DY, double-Y; X, X-shaped molecules; OC, open circular monomer. The arrows indicate the location that corresponds to the asterisk in the lower panel of (E).
