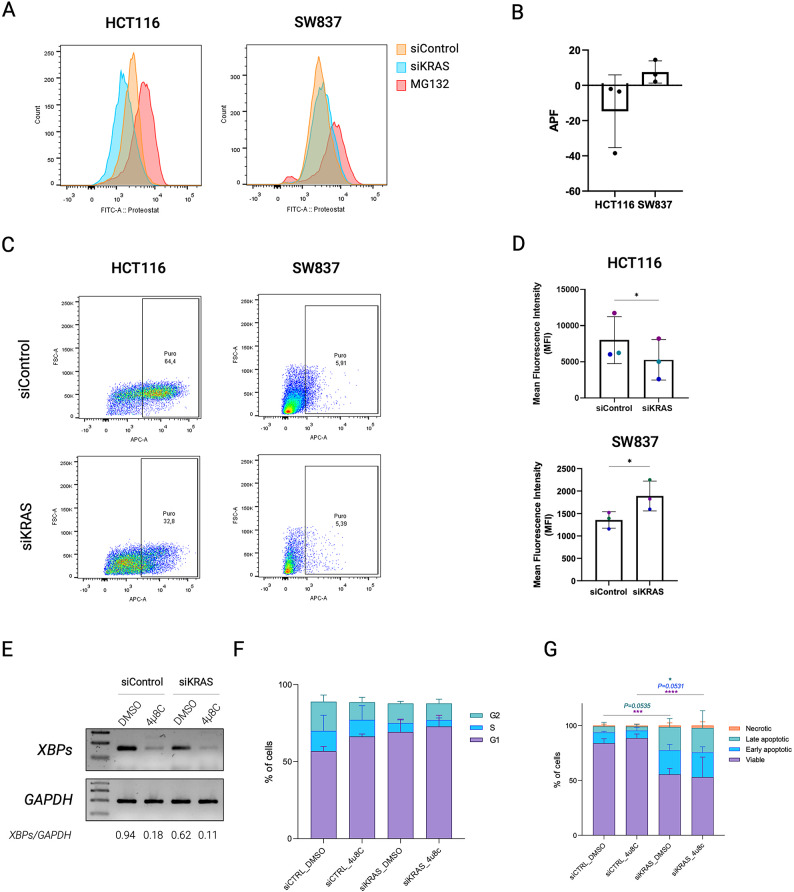
Fig. 6.
Oncogenic KRAS inactivation affects proteostasis. (A) Representative overlaid histograms for the fluorescence intensity of siControl and siKRAS HCT116 and SW837 cells. MG132 was used as positive control. (B) The mean fluorescence intensity values are used to compute the aggresome propensity factor (APF). Data shows the results of three biological replicates and are shown as mean ± SD. (C) Representative flow cytometry profiles for the fluorescence intensity of siControl and siKRAS HCT116 and SW837 cells. (D) The mean fluorescence intensity values of the positive population were used to compute the protein translation potential. Data shows the results of three independent experiments and are shown as mean ± SD. (E) Representative agarose gel of XBPsplicing inhibition after treatment with 4µ8C XBPsplicing and GAPDH PCR products were loaded in different gels. Uncropped versions of the images are provided in Supplementary Fig. 11. (F) Cell cycle analysis in HCT116 cells treated with 4µ8C. Bar graphs illustrate the percentage of cells G0/G1, S, G2/M. (G) Apoptosis analysis of HCT116 cells treated with 4µ8C. Bar graphs represent the percentage of cells that are in viable, early or late apoptosis, and necrotic states. Two-way ANOVA with multiple comparisons was used to compare different populations in siControl with siKRAS conditions (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 00001). All the bar graphs represent the mean ± SD of at least three independent experiments.