Abstract
A comparison of two PCR-based human papillomavirus (HPV) DNA detection and genotyping systems (PGMY LBA and SPF10 LiPA) was conducted in two laboratories. Both systems are based on broad-spectrum PCR for the detection of HPV DNA, followed by reverse hybridization with type-specific probes. A total of 400 selected cervical scrape specimens in PreservCyt solution (55% normal cytology, 18% atypical squamous cells of unknown significance, 14.8% low-grade squamous intraepithelial lesions [SIL], and 12.5% high-grade SIL) were tested for the presence of HPV DNA. In this selected group of specimens, the overall agreement between the two methods for the detection of any HPV DNA was high (κ = 0.859). When the 20 common HPV genotypes identified by both methods were considered (HPV types 6, 11, 16, 18, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 56, 58, 59, 66, and 68), compatible genotype-specific results were observed in 96.5% of the samples, even when multiple HPV genotypes were present. However, for some specific HPV genotypes, there were significant differences in HPV detection by the two methods. PGMY LBA detected more HPV type 42 (P = 0.002), HPV type 56 (P = 0.039), and HPV type 59 (P < 0.001), whereas SPF10 LiPA detected more HPV type 31 (P < 0.001) and HPV type 52 (P = 0.031). For the remaining genotypes, including HPV types 16 and 18, the results obtained by the two methods were not significantly different. In general, both genotyping methods are highly suitable for clinical and epidemiological studies.
Human papillomavirus (HPV) infection is associated with an increased risk for the development of cervical neoplasia (15, 22). Accurate type-specific diagnosis of HPV infections requires sensitive molecular methods, such as PCR. The accurate detection of HPV DNA by PCR is hampered by the existence of a large number of viral genotypes with highly diverse nucleotide sequences (2, 5, 23, 25). PCR-based HPV detection methods have been used for detailed clinical, epidemiological, and natural history studies to elucidate the importance of the different HPV genotypes (7, 10, 11, 21, 26). Among the genotypes occurring in the anogenital region, high-risk and low-risk groups have been identified based on their epidemiological association with the development of cervical cancer (4, 19, 27). Therefore, reliable identification of HPV genotypes, in combination with cytological screening, may be relevant for patient management. In addition, to study the effects of antiviral treatment or type-specific vaccination, accurate HPV genotyping methods are essential for the selection and monitoring of study subjects.
Various PCR-based methods have been described for the identification of HPV genotypes. Individual genotypes can be detected by type-specific PCR primer sets (1, 24). However, these require the performance of multiple parallel assays for each sample, and type-specific PCR primers have not been reported for each HPV genotype. Alternatively, general PCR primer sets can be used, permitting simultaneous amplification of a broad range of HPV genotypes (8, 13, 17, 20). The products of such general amplification reactions can be subsequently analyzed by direct sequencing, restriction fragment length polymorphism, or type-specific probe hybridization. Quality assurance of PCR-based methods in general is a crucial aspect of molecular diagnosis, but data comparing the performance of the reported HPV testing methods are limited (1, 6, 12, 18, 20)
Recently, two independent reverse hybridization assays were developed. The first system, called the line blot assay (LBA), uses a primer set, designated PGMY (8) and based on the MY09/11 primer set, which amplifies a 450-bp fragment within the HPV L1 region (9). The LBA identifies 27 different HPV genotypes. The second system, designated the line probe assay (LiPA), is based on the SPF10 PCR primer set, which amplifies a fragment of only 65 bp within the L1 region. SPF10 amplimers are first tested in a microtiter plate general hybridization assay to detect HPV DNA positivity. Subsequently, the positive samples are analyzed by SPF10 LiPA, which permits the identification of 25 different HPV genotypes (16, 17). Both reverse hybridization systems use type-specific probes selected from the interprimer region of each PCR primer set.
The aim of the present study was to compare both reverse hybridization assays by use of a large number of clinical samples, with a focus on HPV types 16 and 18. Aliquots of clinical materials were exchanged and tested by two laboratories under blinded conditions.
MATERIALS AND METHODS
Samples.
Cervical specimens were obtained from a total of 400 women and were resuspended in approximately 18 ml of PreservCyt solution (Cytyc Corp., Boxborough, Mass.). The panel represented a selection of samples from a wide variety of cytological diagnoses, including normal, atypical squamous cells of unknown significance (ASCUS), and both low- and high-grade abnormalities. Samples were obtained from women attending clinics for routine gynecologic care or during follow-up after treatment.
Overall, the majority of the samples (n = 219; 54.8%) were from women with a normal Pap smear, 72 (18.0%) showed ASCUS, 59 (14.8%) showed low-grade squamous intraepithelial lesions (SIL), and 50 (12.5%) showed high-grade SIL. Each of the two participating laboratories (A and B) selected a panel of 200 samples and prepared duplicate aliquots of 500 μl. Samples were stored at 4°C. Each laboratory transferred a discrete aliquot of each sample to the other laboratory. Thus, a total of 400 samples were tested independently by both laboratories.
Sample preparation.
Each of the laboratories used a different method to isolate DNA from the specimen aliquots. Laboratory A used 500 μl of the original sample, which was centrifuged at 13,000 × g for 15 min. The supernatant was completely removed, and the pellet was dried overnight at room temperature. The dried pellet was resuspended in 50 μl of digestion solution (200 μg of proteinase K per ml, 0.1% Laureth-12, 10 mM Tris-HCl [pH 8.5], 0.1 mM EDTA) and incubated at 56°C for 1 h. The proteinase K was inactivated at 95°C for 15 min. For each amplification reaction, 5 μl of this crude digest was used, representing 50 μl of the original sample. Digests were stored at −20°C.
Laboratory B used 200 μl of the original sample, and DNA was isolated by incubation with guanidinium isothiocyanate followed by capture onto silica particles as described earlier (3). DNA was eluted into 100 μl of 10 mM Tris-HCl [pH 8.3]-0.1 mM EDTA. For each amplification reaction, 10 μl was used, representing 20 μl of the original sample. The purified DNA was stored at −20°C.
PGMY PCR and LBA.
Laboratory A used the PGMY PCR primer set as described earlier (8) but used 5 pmol of each of the β-globin primers GH20 and PC04 instead of 2.5 pmol. PCR products were analyzed with an LBA as described earlier (9), discriminating HPV types 6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 55, 56, 57, 58, 59, 66, 68, 73, 82, 83, and 84. Part of the human β-globin gene was amplified in each sample as a control of specimen adequacy. Appropriate negative and positive controls were used to monitor the performance of the method, and appropriate cell-based controls were used to monitor contamination and assay performance.
SPF10 PCR and LiPA.
Laboratory B used the SPF10 PCR primer set as described earlier (17). Amplification products were first tested by probe hybridization in a microtiter plate assay to detect the presence of HPV DNA. Amplicons from HPV-positive samples were subsequently analyzed by reverse hybridization by the LiPA, which permits specific detection of 25 HPV genotypes, i.e., HPV types 6, 11, 16, 18, 31, 33, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 66, 68, 70, 73, and 74 (16). Part of the human β-globin gene was amplified in each sample as a control of specimen adequacy. Appropriate negative and positive controls were used to monitor the performance of the method.
Type-specific PCR for HPV types 16 and 18.
Type-specific detection of HPV types 16 and 18 was performed in an independent third laboratory with type-specific PCR primers for each genotype as described earlier (1). The HPV type 16 PCR primers generate a fragment of 96 bp from the E6-E7 region, and the HPV type 18 PCR primers generate a 115-bp fragment from the L1 region.
Data analysis and statistics.
All data were analyzed using SPSS for Windows, version 8.0, and McNemar's, chi-square, and Cohen's kappa tests. For comparison, only the 20 common HPV genotypes detected by both methods (HPV types 6, 11, 16, 18, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 56, 58, 59, 66, and 68) were considered. Genotyping results were considered concordant when analysis of the specimens by the two methods yielded completely identical (single or multiple) genotypes. Results were classified as compatible when multiple genotypes were detected and at least one genotype was found identical by both assays. When completely different genotypes were detected, results were classified as discordant.
RESULTS
A total of 400 selected cervical samples in PreservCyt solution were tested in two laboratories using the PGMY LBA and SPF10 LiPA systems. Using the PGMY LBA, laboratory A detected HPV DNA in 255 samples (63.8%). Using the SPF10 LiPA, laboratory B detected HPV DNA in 257 samples (64.3%). There was agreement for HPV DNA detection by both methods in 374 (93.5%) of the 400 samples tested (κ = 0.859; 95% confidence interval, 0.807 to 0.911), and HPV detection was not significantly different (McNemar's test: P = 0.69). The human β-globin gene target was detected in 399 of the 400 samples. One sample remained β-globin negative at laboratory A.
HPV DNA-positive samples were subsequently genotyped by the different reverse hybridization methods, LBA and LiPA. The majority of the HPV DNA-positive samples contained a single HPV genotype. However, in a considerable proportion of the samples, multiple HPV genotypes were identified. The results of each HPV DNA detection method are summarized in Table 1. When all samples with multiple genotypes were combined, HPV detection by the two methods was not significantly different (chi-square test: P = 0.942).
TABLE 1.
Identification of single and multiple genotypes by PGMY LBA and SPF10 LiPAa
| Genotype(s) | No. (%) of samples found positive by:
|
|
|---|---|---|
| PGMY LBA (n = 255) | SPF10 LiPA (n = 257) | |
| Single | 140 (54.9) | 152 (59.3) |
| Multiple | 115 (45.1) | 105 (40.7) |
| 2 | 64 (25.1) | 68 (26.5) |
| 3 | 25 (9.8) | 27 (10.5) |
| 4 | 16 (6.3) | 6 (2.3) |
| >4 | 10 (3.9) | 4 (1.6) |
The difference between the two methods for the detection of single versus multiple genotypes (total combined categories of all samples containing more than one genotype) was not significant (chi-square test: P = 0.942).
As stated in Materials and Methods, genotyping results were scored as concordant, compatible, and discordant. Since the two reverse hybridization methods use probes for different but overlapping ranges of HPV genotypes, only the genotypes detected by both systems (i.e., HPV types 6, 11, 16, 18, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 56, 58, 59, 66, and 68) were considered for comparison. The results are summarized in Table 2. Of the 243 samples where HPV DNA was detected by both methods, 15 samples (6.2%) were excluded from the analysis, because the HPV genotypes were not identified by either SPF10 LiPA or PGMY LBA. Among the remaining 228 HPV DNA-positive samples, 128 (56.1%) contained concordant genotypes, 92 (40.4%) contained compatible genotypes, and 8 (3.5%) contained discordant genotypes. Thus, the two reverse hybridization methods revealed either identical or compatible genotypes in 96.5% of the samples. The typing results were compatible in 92 samples (i.e., both methods identified at least one identical genotype), and these were further analyzed. In 43 of these 92 samples, PGMY LBA identified additional types compared to SPF10 LiPA. Conversely, SPF10 LiPA identified additional types in 35 samples compared to PGMY LBA. The difference in the detection of additional genotypes by the assays was not significant (chi-square test: P = 0.233). In 14 samples, both PGMY LBA and SPF10 LiPA identified extra genotypes.
TABLE 2.
Agreement between HPV genotypes detected by both PGMY LBA and SPF10 LiPAa
| Type | % Agreement for:
|
||||
|---|---|---|---|---|---|
| PGMY LBA
|
SPF10 LiPA
|
Total (n = 228) | |||
| Single types | Multiple types | Single types | Multiple types | ||
| Concordant | 96 | 32 | 96 | 32 | 128 |
| Compatible | 24 | 68 | 26 | 66 | 92 |
| PGMY LBA additional types | 0 | 43 | 26 | 17 | 43 |
| SPF10 LiPA additional types | 24 | 11 | 0 | 35 | 35 |
| Both additional types | 0 | 14 | 0 | 14 | 14 |
| Discordant | 7 | 1 | 8 | 0 | 8 |
HPV types 6, 11, 16, 18, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 56, 58, 59, 66, and 68.
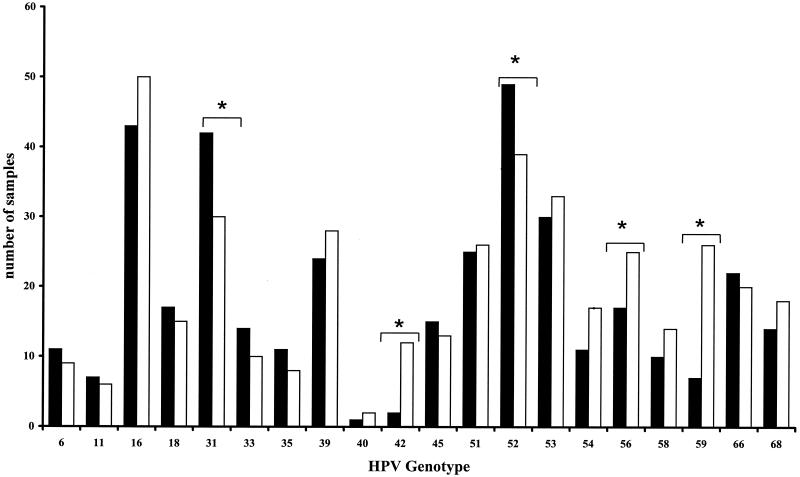
Subsequently, a comparison of the specific identification of individual HPV genotypes in this selected group of specimens by each of the two methods was made, and the results are summarized in Fig. 1 and Table 3. For most genotypes, including HPV types 16 and 18, the results obtained by the two methods were not significantly different. However, PGMY LBA was more sensitive for HPV types 42, 56, and 59, whereas SPF10 LiPA was more sensitive for HPV types 31 and 52.
FIG. 1.
Detection of different HPV genotypes by two different general PCR reverse hybridization methods. Results are shown for HPV genotypes that can be identified by both systems (HPV types 6, 11, 16, 18, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 56, 58, 59, 66, and 68). Closed bars represent results obtained by SPF10 LiPA. Open bars represent results obtained by PGMY LBA. Significant differences are indicated by asterisks (McNemar's test). For the remaining genotypes, the detection rates were not significantly different.
TABLE 3.
Kappa values for HPV genotypes detected by both PGMY LBA and SPF10 LiPAa
| HPV type | No. of samples found positive by:
|
Kappa value (95% confidence interval) | ||
|---|---|---|---|---|
| PGMY LBA | SPF10 LiPA | Both PGMY LBA and SPF10 LiPA | ||
| 6 | 9 | 11 | 7 | 0.692 (0.459-0.926) |
| 11 | 6 | 7 | 5 | 0.765 (0.508-1.023) |
| 16 | 50 | 43 | 39 | 0.816 (0.728-0.907) |
| 18 | 15 | 17 | 12 | 0.710 (0.566-0.913) |
| 31 | 30 | 42 | 30 | 0.817 (0.717-0.917) |
| 33 | 10 | 14 | 9 | 0.742 (0.544-0.940) |
| 35 | 8 | 11 | 6 | 0.623 (0.365-0.881) |
| 39 | 28 | 24 | 22 | 0.836 (0.724-0.947) |
| 40 | 2 | 1 | 1 | 0.666 (0.044-1.283) |
| 42 | 12 | 2 | 2 | 0.280 (−0.026-0.585) |
| 45 | 13 | 15 | 11 | 0.779 (0.606-0.950) |
| 51 | 26 | 25 | 20 | 0.770 (0.638-0.901) |
| 52 | 39 | 49 | 35 | 0.772 (0.671-0.873) |
| 53 | 33 | 30 | 29 | 0.914 (0.834-0.989) |
| 54 | 17 | 11 | 9 | 0.631 (0.419-0.842) |
| 56 | 25 | 17 | 15 | 0.699 (0.539-0.860) |
| 58 | 14 | 10 | 10 | 0.828 (0.663-0.993) |
| 59 | 26 | 7 | 7 | 0.408 (0.199-0.617) |
| 66 | 20 | 22 | 18 | 0.849 (0.731-0.968) |
| 68 | 18 | 14 | 12 | 0.740 (0.567-0.913) |
HPV types 6, 11, 16, 18, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 56, 58, 59, 66, and 68.
To assess the accurate detection and identification of HPV types 16 and 18, a discrepancy analysis using type-specific PCR primers was performed on a subset of samples (n = 40). This subset included samples that were discrepant between the PGMY LBA and SPF10 LiPA assays as well as concordant samples as controls. To determine the influence of sampling errors and differences between pre-PCR sample treatments, pretreated samples from both laboratories were used for type-specific PCR.
Twenty-six samples were evaluated by HPV type 16-specific PCR (1). Overall, the three methods yielded concordant results in 16 samples, but in 10 samples discrepancies remained. Fourteen samples were tested for HPV 18 by type-specific PCR (1). Overall, results were concordant among the three methods in 11 samples, but discrepancies remained in 3 samples.
DISCUSSION
Detection and identification of the broad range of clinically relevant HPV genotypes require reliable molecular tools to permit adequate diagnosis for various clinical samples, such as cervical and biopsy specimens. In the present study, we tested a total of 400 cervical scraping specimens for the presence of HPV DNA by two different methods.
The test panel was selected to contain a spectrum of different HPV genotypes and cannot be considered representative of the prevalence of HPV in a random population. Furthermore, each laboratory used a different sample preparation method and included a different volume of the original specimen in the PCR assay. Therefore, the composition of the panel of specimens in the present study does not fully address the sensitivity of the two PCR methods; rather, the analysis was focused on a comparison of HPV genotyping by the two reverse hybridization methods, in particular for HPV types 16 and 18.
Cervical scraping samples the surface of the cervical epithelium, and the number of cells obtained is variable. The cells from the cervical specimens reported in the present study were placed in PreservCyt solution. This specimen medium permits more reliable cytological examinations and may reduce sampling errors between different liquid cytology medium aliquots from a single cervical swab specimen. The potential for sample heterogeneity is an important factor to consider, since aliquots of each sample were tested by two different methods in separate laboratories.
The two molecular methods used different PCR primer sets. Both sets permit amplification of a broad range of HPV genotypes, and both target the L1 region of the HPV genome; however, there are several important differences (8, 17). In general, the efficacy of a general PCR primer set will depend on the size of the amplimer, the degeneracy or number and spectrum of the PCR primers used, the multiple HPV genotypes present within individual specimens, and sequence variations at the priming sites in the different HPV genotypes (20).
The PGMY primer set comprises a pool of 18 different primers (5 forward and 13 reverse) and amplifies a fragment of approximately 450 bp at an annealing temperature of 55°C. The SPF10 primer set comprises a total of 10 different PCR primers (6 forward and 4 reverse) and amplifies a fragment of only 65 bp at an annealing temperature of 52°C. The forward SPF10 priming site overlaps the forward priming site of the PGMY primer set. Both sets comprise distinct primers without degeneracies. Primers with degenerate base sequences are not suitable for standardization, since the insertion of nucleotides at degenerate positions during the synthesis of an oligonucleotide is a random and irreproducible process (8). Therefore, mixtures of defined primers without degenerate positions have been developed, resulting in higher reproducibility and more robust assays. The SPF10 PCR primers contain an inosine nucleotide at some positions to optimize the match with the primer target regions of various HPV genotypes and to reduce the number of primers required for general HPV DNA detection (16). In the present study, the two PCR methods showed different sensitivities for a number of HPV genotypes. The PGMY primer set was more sensitive for HPV types 42, 56, and 59, but the SPF10 primer set was more sensitive for HPV types 31 and 52. For the remaining genotypes, there were no significant differences in HPV detection. These type-specific effects can at least be partly attributed to an insufficient match between any of the primers in the mixture and the primer target sites of some HPV types.
When multiple HPV genotypes are present in a sample, the situation becomes more complex. Besides the preferential use of a type-specific subset of PCR primers, competition between the different HPV genotypes may play an important role. If two HPV genotypes are present in equimolar amounts, it is likely that both will be amplified and identified. However, if one genotype is present in great molar excess over the other, it becomes more likely that the minor genotype will be outcompeted and remain below the detection limit of the assay. This phenomenon is illustrated in our study by two findings. First, the HPV typing results for a considerable number of samples that contained multiple genotypes were not completely identical between the two methods but were scored as having compatible genotypes (i.e., at least one identical genotype was identified by both methods). This result is partly due to the fact that each PCR primer set has a slightly different preference for the specific genotypes in the sample and will preferentially amplify subsets of genotypes. Second, we also tested a subset of samples that yielded discrepant HPV type 16 or HPV type 18 results in the reverse hybridization assays by a type-specific PCR for HPV type 16 or HPV type 18. The fact that aliquots from laboratories A and B yielded different results in the type-specific PCR illustrates the important effect of sampling errors, even when aliquots are derived from the same homogenized sample. Presumably, such sampling errors are especially important when HPV is present at very low concentrations. HPV type 16 or HPV type 18 was detected in some samples by the type-specific PCR but remained undetected by both PCR primer sets. In virtually all of these discrepant samples, other HPV genotypes had been identified, indicating the presence of multiple types. Thus, if HPV type 16 or HPV type 18 alone is present at low concentrations, it will most likely be adequately detected by both PCR primer sets. However, if HPV type 16 or HPV type 18 is present at low concentrations among high concentrations of other genotypes, these PCR assays may yield a false-negative result for HPV type 16 or HPV type 18. In fact, this scenario may be true for all genotypes present at very low concentrations among other genotypes. Thus, the prevalence of infections with multiple HPV genotypes is likely to be underestimated by a PCR primer set designed to target a broad spectrum of HPV genotypes. If one is interested in the diagnosis of a particular HPV genotype, type-specific PCR may be more suitable than broad-spectrum general PCR (12, 14).
In conclusion, the present study demonstrates that the genotyping results obtained by PGMY LBA and SPF10 LiPA are highly comparable, with a very small number of discordant results. These findings confirm that both reverse hybridization methods are useful for HPV detection and permit accurate identification of a broad range of HPV genotypes.
REFERENCES
- 1.Baay, M. F., W. G. Quint, J. Koudstaal, H. Hollema, J. M. Duk, M. P. Burger, E. Stolz, and P. Herbrink. 1996. Comprehensive study of several general and type-specific primer pairs for detection of human papillomavirus DNA by PCR in paraffin-embedded cervical carcinomas. J. Clin. Microbiol. 34:745-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard, H. U., S. Y. Chan, M. M. Manos, C. K. Ong, L. L. Villa, H. Delius, C. L. Peyton, H. M. Bauer, and C. M. Wheeler. 1994. Identification and assessment of known and novel human papillomaviruses by polymerase chain reaction amplification, restriction fragment length polymorphisms, nucleotide sequence, and phylogenetic algorithms. J. Infect. Dis. 170:1077-1085. [DOI] [PubMed] [Google Scholar]
- 3.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosch, F. X., M. M. Manos, N. Munoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, K. V. Shah, et al. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl. Cancer Inst. 87:796-802. [DOI] [PubMed] [Google Scholar]
- 5.Chan, S. Y., H. Delius, A. L. Halpern, and H. U. Bernard. 1995. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J. Virol. 69:3074-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniel, R. W., L. Ahdieh, D. Hayden, S. Cu-Uvin, and K. V. Shah. 2000. Intra-laboratory reproducibility of human papillomavirus identification in cervical specimens by a polymerase chain reaction-based assay. J. Clin. Virol. 19:187-193. [DOI] [PubMed] [Google Scholar]
- 7.Elfgren, K., M. Kalantari, B. Moberger, B. Hagmar, and J. Dillner. 2000. A population-based five-year follow-up study of cervical human papillomavirus infection. Am. J. Obstet. Gynecol. 183:561-567. [DOI] [PubMed] [Google Scholar]
- 8.Gravitt, P. E., C. L. Peyton, T. Q. Alessi, C. M. Wheeler, F. Coutlée, A. Hildesheim, M. Schiffman, D. R. Scott, and R. J. Apple. 2000. Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 38:357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gravitt, P. E., C. L. Peyton, R. J. Apple, and C. M. Wheeler. 1998. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J. Clin. Microbiol. 36:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hildesheim, A., M. H. Schiffman, P. E. Gravitt, A. G. Glass, C. E. Greer, T. Zhang, D. R. Scott, B. B. Rush, P. Lawler, M. E. Sherman, R. J. Kurman, and M. M. Manos. 1994. Persistence of type-specific human papillomavirus infection among cytologically normal women. J. Infect. Dis. 169:235-240. [DOI] [PubMed] [Google Scholar]
- 11.Ho, G. Y., R. Bierman, L. Beardsley, C. J. Chang, and R. D. Burk. 1998. Natural history of cervicovaginal papillomavirus infection in young women. N. Engl. J. Med. 338:423-428. [DOI] [PubMed] [Google Scholar]
- 12.Husnjak, K., M. Grce, L. Magdic, and K. Pavelic. 2000. Comparison of five different polymerase chain reaction methods for detection of human papillomavirus in cervical cell specimens. J. Virol. Methods 88:125-134. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs, M. V., P. J. Snijders, A. J. van den Brule, T. J. Helmerhorst, C. J. Meijer, and J. M. Walboomers. 1997. A general primer GP5+/GP6(+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 35:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsen, F., M. Kalantari, A. Jenkins, E. Pettersen, G. Kristensen, R. Holm, B. Johansson, and B. Hagmar. 1996. Use of multiple PCR primer sets for optimal detection of human papillomavirus. J. Clin. Microbiol. 34:2095-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kjaer, S. K., A. J. van den Brule, J. E. Bock, P. A. Poll, G. Engholm, M. E. Sherman, J. M. Walboomers, and C. J. Meijer. 1996. Human papillomavirus—the most significant risk determinant of cervical intraepithelial neoplasia. Int. J. Cancer 65:601-606. [DOI] [PubMed] [Google Scholar]
- 16.Kleter, B., L. J. van Doorn, L. Schrauwen, A. Molijn, S. Sastrowijoto, J. ter Schegget, J. Lindeman, B. ter Harmsel, and W. G. V. Quint. 1999. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J. Clin. Microbiol. 37:2508-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleter, B., L. J. van Doorn, J. ter Schegget, L. Schrauwen, C. van Krimpen, M. P. Burger, B. ter Harmsel, and W. G. V. Quint. 1998. A novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am. J. Pathol. 153:1731-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nindl, I., M. Jacobs, J. M. Walboomers, C. J. Meijer, H. Pfister, U. Wieland, T. Meyer, E. Stockfleth, R. Klaes, D. M. von Knebel, A. Schneider, and M. Duerst. 1999. Interlaboratory agreement of different human papillomavirus DNA detection and typing assays in cervical scrapes. Int. J. Cancer 81:666-668. [DOI] [PubMed] [Google Scholar]
- 19.Nobbenhuis, M. A., J. M. Walboomers, T. J. Helmerhorst, L. Rozendaal, A. J. Remmink, E. K. Risse, H. C. van der Linden, F. J. Voorhorst, P. Kenemans, and C. Meijer. 1999. Relation of human papillomavirus status to cervical lesions and consequences for cervical-cancer screening: a prospective study. Lancet 354:20-25. [DOI] [PubMed] [Google Scholar]
- 20.Qu, W., G. Jiang, Y. Cruz, C. J. Chang, G. Y. Ho, R. S. Klein, and R. D. Burk. 1997. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J. Clin. Microbiol. 35:1304-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rozendaal, L., J. Westerga, J. C. van der Linden, J. M. Walboomers, F. J. Voorhorst, E. K. Risse, M. E. Boon, and C. J. Meijer. 2000. PCR based high risk HPV testing is superior to neural network based screening for predicting incident CIN III in women with normal cytology and borderline changes. J. Clin. Pathol. 53:606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schiffman, M., H. M. Bauer, R. Hoover, A. G. Glass, D. M. Cadell, B. B. Rush, D. R. Scott, M. Sherman, R. Kurman, S. Wacholder, C. K. Stanton, and M. M. Manos. 1993. Epidemiologic evidence showing that HPV infection causes most cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 85:958-964. [DOI] [PubMed] [Google Scholar]
- 23.Stewart, A. C., A. M. Eriksson, M. M. Manos, N. Munoz, F. X. Bosch, J. Peto, and C. M. Wheeler. 1996. Intratype variation in 12 human papillomavirus types: a worldwide perspective. J. Virol. 70:3127-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 25.Yamada, T., M. M. Manos, J. Peto, C. E. Greer, N. Munoz, F. X. Bosch, and C. M. Wheeler. 1997. Human papillomavirus type 16 sequence variation in cervical cancers: a worldwide perspective. J. Virol. 71:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ylitalo, N., A. Josefsson, M. Melbye, P. Sorensen, M. Frisch, P. K. Andersen, P. Sparen, M. Gustafsson, P. Magnusson, J. Ponten, U. Gyllensten, and H. O. Adami. 2000. A prospective study showing long-term infection with human papillomavirus 16 before the development of cervical carcinoma in situ. Cancer Res. 60:6027-6032. [PubMed] [Google Scholar]
- 27.zur Hausen, H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]