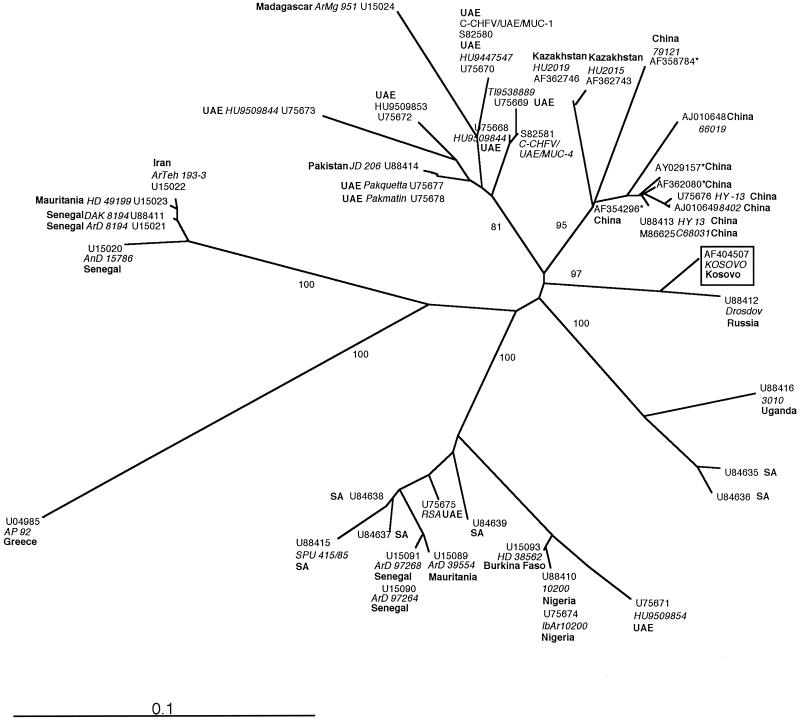
FIG. 1.
Phylogenetic analysis of 46 partial sequences (219 bp) of the S segment of CCHF virus. The sequences were aligned and analyzed with the Phylip 3.57 software package (J. Felsenstein, University of Washington), using the neighbour-joining and maximum-likelihood (results not shown) algorithms. Both methods identified the same seven major genetic groups. The bootstrap values (percentages of 100 replicates) for each group are depicted at the respective branches. The geographic origin (boldface type), the GenBank accession number, and the additional strain denomination, if available (in italics) are given at the terminal branches for each isolate. Strains from the Middle and Far East and from different African regions cluster in clearly separated groups. Higher phylogenetic diversity among strains from the United Arabian Emirates (UAE) is attributable to importation of CCHF virus-infected livestock and ticks from African and Middle Eastern countries (6). SA, South Africa.