Table II.
qRT-PCR and ATH1 expression levels of traditional and novel reference genes
Arabidopsis Gene IDs of traditional reference genes are in bold. % P Calls, Percentage of Present calls within the experimental series, as attributed by MAS5 software.
| Arabidopsis Gene ID | Annotation | Mean ΔCTa | Mean 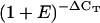
|
Relative Expression | se (n = 20) | ATH1 Absolute/Relative Mean Expressionb | % P Callsb |
|---|---|---|---|---|---|---|---|
| ×106 | |||||||
| AT4G05320 | UBQ10 | 8.76 | 4,270 | 1 | 6.9 E-04 | 6,811/0.412 | 100 |
| AT5G60390 | EF-1α | 9.07 | 3,660 | 0.858 | 6.7 E-04 | 16,528/1 | 100 |
| AT1G13440 | GAPDH | 9.50 | 2,488 | 0.583 | 3.5 E-04 | 8,220/0.497 | 100 |
| AT4G27960 | UBC9 | 10.98 | 1,160 | 0.272 | 1.5 E-04 | 9,979/0.604 | 100 |
| AT3G18780 | ACT2 | 11.74 | 917 | 0.215 | 1.9 E-04 | 5,347/0.323 | 99 |
| AT5G46630 | Clathrin adaptor complex subunit | 12.71 | 421 | 0.099 | 3.9 E-05 | 1,115/0.068 | 100 |
| AT5G08290 | YLS8 | 13.60 | 343 | 0.081 | 3.9 E-05 | 2,924/0.177 | 100 |
| AT4G33380 | Expressed | 13.45 | 342 | 0.080 | 3.2 E-05 | 310/0.019 | 100 |
| AT2G32170 | Expressed | 14.47 | 246 | 0.058 | 3.3 E-05 | 337/0.020 | 100 |
| AT4G34270 | TIP41-like | 13.68 | 243 | 0.057 | 2.9 E-05 | 353/0.021 | 90 |
| AT5G25760 | UBC | 13.82 | 190 | 0.044 | 2.6 E-05 | 422/0.026 | 100 |
| AT1G13320 | PDF2 | 13.41 | 189 | 0.044 | 2.7 E-05 | 713/0.043 | 100 |
| AT2G28390 | SAND family | 14.69 | 167 | 0.039 | 1.9 E-05 | 415/0.025 | 99 |
| AT4G26410 | Expressed | 14.11 | 132 | 0.031 | 1.7 E-05 | 617/0.037 | 100 |
| AT3G01150 | PTB | 18.56 | 40 | 0.009 | 4.7 E-06 | 247/0.015 | 99 |
| AT5G55840 | PPR repeat | 18.59 | 38 | 0.009 | 4.4 E-06 | 24.8/0.001 | 99 |
| AT1G58050 | Helicase | 16.81 | 36 | 0.008 | 3.9 E-06 | 179/0.011 | 97 |
| AT3G53090 | UPL7 | 19.70 | 22 | 0.005 | 2.1 E-06 | 112/0.007 | 100 |
| AT5G15710 | F-box family | 17.53 | 15 | 0.003 | 1.9 E-06 | 99.8/0.006 | 98 |
| AT4G38070 | bHLH | 22.46 | 6 | 0.001 | 1.1 E-06 | 14.1/0.0009 | 84 |
| AT5G12240 | Expressed | 21.91 | 1.6 | 0.00038 | 1.8 E-07 | 5.3/0.0003 | 90 |
| AT1G62930 | PPR repeat | 24.02 | 0.4 | 0.00009 | 6.3 E-08 | 95.1/0.006 | 99 |
| AT3G32260 | Hypothetical | NA | NA | NA | NA | 14.0/0.0008 | 88 |
| AT2G07190 | Hypothetical | NA | NA | NA | NA | 10.7/0.0006 | 89 |
| AT1G47770 | Hypothetical | NA | NA | NA | NA | 5.6/0.0003 | 59* |
Difference in threshold cycle number relative to LjLb2.
Based on the developmental series; NA, data not available since no PCR amplification could be obtained. *, Chosen based on its presence in the Top100 lists of the shoot and root abiotic stress and nutrient stress series and because it showed >80% Present calls in all three.
