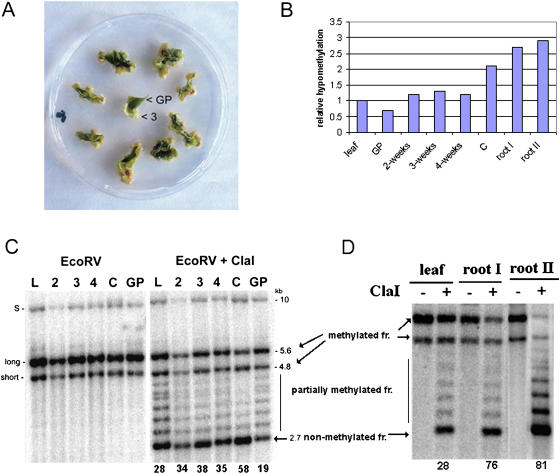
Figure 2.
Hypomethylation of rDNA units in dedifferentiated callus and root tip cells. A, A single leaf cut into several pieces was laid onto callus induction medium. At 2-, 3-, and 4-week intervals, the dedifferentiated cells formed at the edges of green tissue were collected and DNA extracted. The nondedifferentiated green tissue (GP) was analyzed after 3 weeks of cultivation. “3”, 3-week callus tissue from which DNA was extracted. B, Graph showing relative hypomethylation of rDNA units in tobacco tissues. The data are taken from hybridization experiments in C and D. The hypomethylation is expressed as the ratio of signal in nonmethylated fraction (2.7 kb) to total signal (%). The value obtained for leaf was arbitrarily chosen as 1. Abbreviations are as in C. C, Methylation changes in rDNA units during callus formation. DNAs digested with EcoRV alone or in combination with methylation-sensitive ClaI are shown. Southern-blot hybridization of restricted genomic DNA was carried out with the 18S rDNA probe to reveal methylation of the IGS SR-VI subregion. The 5.6- and 4.8-kb EcoRV/ClaI hybridization bands represent molecules with fully methylated A1/A2 subrepeats; a ladder of fragments in the 3.1- to 4.8-kb region represents partially methylated molecules. The 2.7-kb band (arrow) is indicative of demethylated ClaI sites proximal to the18S gene. Note that the 2.7-kb band becomes stronger in the course of callus proliferation. DNA from the green part of nondedifferentiating tissue was slightly less digested with ClaI than the original leaf. L, Original leaf; 2, 3, 4, dedifferentiating tissue 2, 3, and 4 weeks, respectively, after passage of the leaf explants onto callus-inducing medium; C, fully dedifferentiated callus; GP, green tissue; S, rDNA family derived from N. sylvestris; long, short, rDNA families derived from N. tomentosiformis. Numbers below lanes indicate intensities of hybridization signals in nonmethylated 2.7-kb fractions as a percentage of total signal in lanes. D, Methylation analysis of root and leaf rDNA. Southern-blot hybridization was carried out as in C. DNAs of leaf, 10-mm (I), and 3-mm root tips (II) were isolated from a single plant. Quantification of signals was carried out as in C.