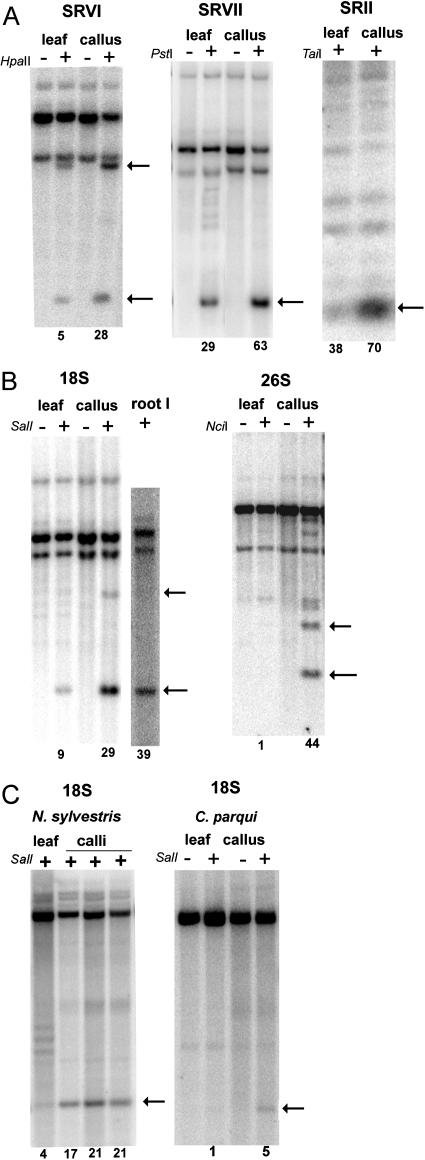
Figure 3.
Southern-blot hybridization analysis of cytosine methylation in fully dedifferentiated callus and parental leaf of N. tabacum (A and B), N. sylvestris, and C. parqui (C). DNAs were predigested with methylation-insensitive restriction enzymes (Table I) to delimit intergenic (A) and genic (B and C) subregions of the rDNA cistron (Fig. 1). The digestion with methylation-sensitive enzyme is indicated with a plus (+). HpaII was used in combination with MboI methylation-insensitive restriction endonuclease; PstI and SalI with EcoRV; TaiI and NciI with BstNI. Arrows indicate fragments corresponding to molecules with nonmethylated restriction sites. The signals in bands were quantified by a phosphor imager and the relative proportion of units with nonmethylated sites is shown below the lanes.