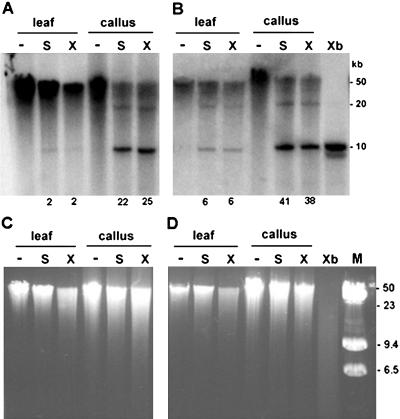
Figure 4.
Large-scale organization of hypomethylated units. The DNA of approximately 50 kb was digested with methylation-sensitive SalI (S lines) and XhoI (X lines) enzymes, separated in 0.6% agarose gel, and hybridized with the 18S rDNA probe. Digestion with CG methylation-insensitive XbaI enzyme was used to reveal a repeat monomer of approximately 10 kb (Xb line). Note that callus DNAs from both Vielblattriger (A) and SR-1 (B) varieties contain significant hybridization signal in a monomeric fraction. Minus (−) indicates nondigested DNA. Numbers below lanes indicate intensities of hybridization signals in nonmethylated, approximately 10-kb fractions as a percentage of total signal in lanes (6–50 kb). Ethidium bromide gels prior to DNA transfer are shown in C and D to reveal global genome methylation at respective restriction sites. C and D correspond to blots A and B, respectively. M, Size markers (lambda + lambda/HindIII).