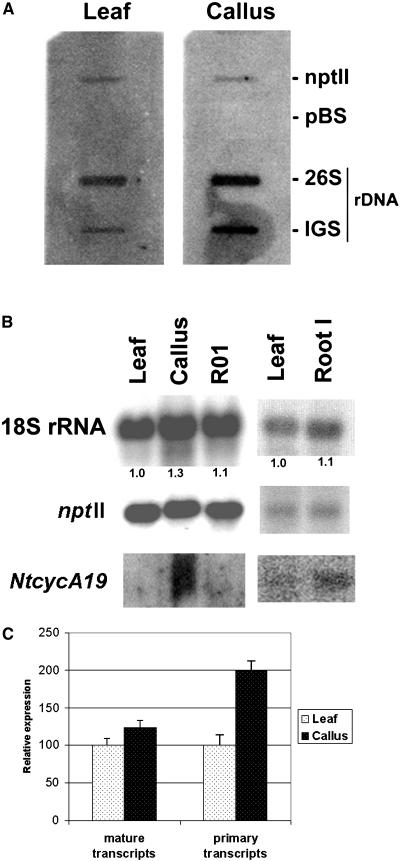
Figure 6.
Expression analysis in parental leaves, derived calli, and regenerated plants. A, Representative slot-blot hybridization with nascent RNA synthesized by run-on transcription using nuclei from the parental leaf and derived callus. The slots contained plasmids carrying the 26S gene and the A1/A2 subrepeat (IGS) as inserts. The expression of nonsilenced and nonmethylated nptII transgene served as a positive control. The plasmid vector (pBluescript; Stratagene) hybridization was used as a negative control (pBS). B, Representative northern-blot hybridization of steady-state transcripts. Total RNAs were isolated from different tissues and were subsequently hybridized on blots with the nptII, 18S rRNA, and NtcycA19 gene probes. Numbers below the lanes indicate the multiple increase in 18S/nptII ratio. For leaf, the ratio was arbitrarily chosen as 1. R01, Tissue culture regenerant; root I, RNA sample from an approximately 10-mm root tip. C, Quantification of the primary and steady-state (mature transcript) levels. The signals in A and B were quantified using a phosphor imager. The data from two run-on assays and six northern-blot experiments, independently conducted, were collected, normalized to the nptII transgene expression, and expressed as an average.