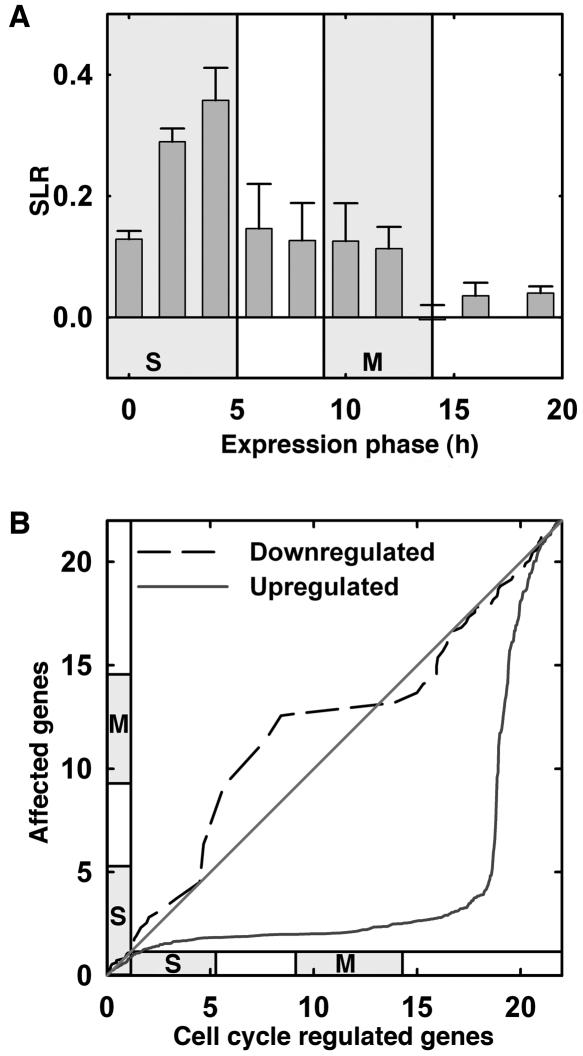
Figure 2.
Increased expression of cell-cycle-phase-specific genes in E2Fa-DPaOE plants. Expression peaks for 9,910 genes during the 22-h cell cycle of Arabidopsis (Menges et al., 2003) were used. In this experiment, transcript levels were measured at 10 time points. The first time point (0 h) represents cells blocked in early S phase. Synchronized cells completed S phase at 5 h and went through mitosis from 9–14 h after release from the block. Identification of 9,910 genes that were differentially expressed during the cell cycle and determination of their expression peaks were described before (Menges et al., 2003). A, The 9,910 genes identified by Menges et al. (2003) were sorted into 10 bins, which represent the 10 time points of measurements, based on their maximal expression during the cell cycle. Shown are the mean ± se of the SLRs (logarithms to base 2) between expression signals in wild-type and E2Fa-DPaOE plants for all genes in a bin. Note that the 8th bin, left to the time point of 15 h, holds only a small number of genes. Shaded areas represent periods of S phase and mitosis, respectively. B, The q-q plots for genes significantly modified in E2Fa-DPaOE seedlings versus all cell-cycle-regulated genes. Slopes close to 0 indicate an overrepresentation of the number of E2Fa-DPa-affected genes among the number of cell-cycle-regulated genes in the time window indicated on the y axis.