Table 2.
The protease enzyme residues that were seen in interaction with the hit compounds and control molecule in the following table.
| S.No | Compound ID | Chemical Structure | Binding affinity |
|---|---|---|---|
| 1. |
CMNPD229 (3aR,4aR,8aS,9 S)-1-isopropyl-3a,8a-dimethyl-5-methylene-1,2,3,3a,4,4a,5,6,7,8,8a,9-dodecahydrobenzo[f]azulene-1,4a,9-triol |
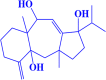
|
-8.1 kcal/mol |
| 2. |
ZINC000000018635 3-(methylamino)-2,3,4,9-tetrahydro-1 H-carbazole-6-carboxamide |
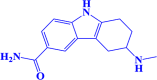
|
-8 kcal/mol |
| 3. |
ZINC000005425464 2-amino-6-(benzyloxy)-9 H-purine-1,3,7-triium |

|
-7.8 kcal/mol |
| 4. |
CMNPD450 2-(11-methyl-3,7-dimethylenedodec-11-en-1-yl)benzene-1,4-diol |
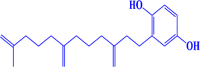
|
-7.7 kcal/mol |
| 5. |
CMNPD1188 8-ethyl-3-hydroxy-3,5b,8,11a,13a-pentamethyl-13-oxoicosahydrochryseno[1,2-c]furan-4-yl acetate |
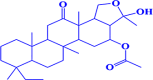
|
-7.6 kcal/mol |
| 6. |
CMNPD2030 3b-hydroxy-3,3,7a-trimethyl-4-methylenedecahydro-1 H-cyclopenta[a]pentalene-1,5-diyl diacetate |

|
-7.3 kcal/mol |
| 7. |
ZINC000000005109 6-(bicyclo[2.2.1]heptan-2-ylamino)-9-methyl-2,3,7,8-tetrahydro-1 H-purine-1,3,9-triium |
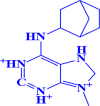
|
-7.3 kcal/mol |
| 8. |
ZINC000100004253 (E)-5-amino-11-ethylidene-7-methylene-5,6,7,8,9,10-hexahydro-5,9-methanocycloocta[b]pyridin-2-ol |
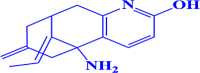
|
-7.3 kcal/mol |
| 9. |
ZINC000002525891 3-(1-hydroxy-2-((methylamino)methyl)cyclohexyl)phenol |
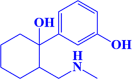
|
-7.2 kcal/mol |
| 10. |
ZINC000038810990 1-(2-(1-carboxyvinyl)hydrazinyl)phthalazine-2,3-diium |
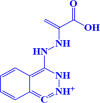
|
-7.1 kcal/mol |
| 11. |
Control Chloroquine |
|
-6.8 Kcal/mol |
