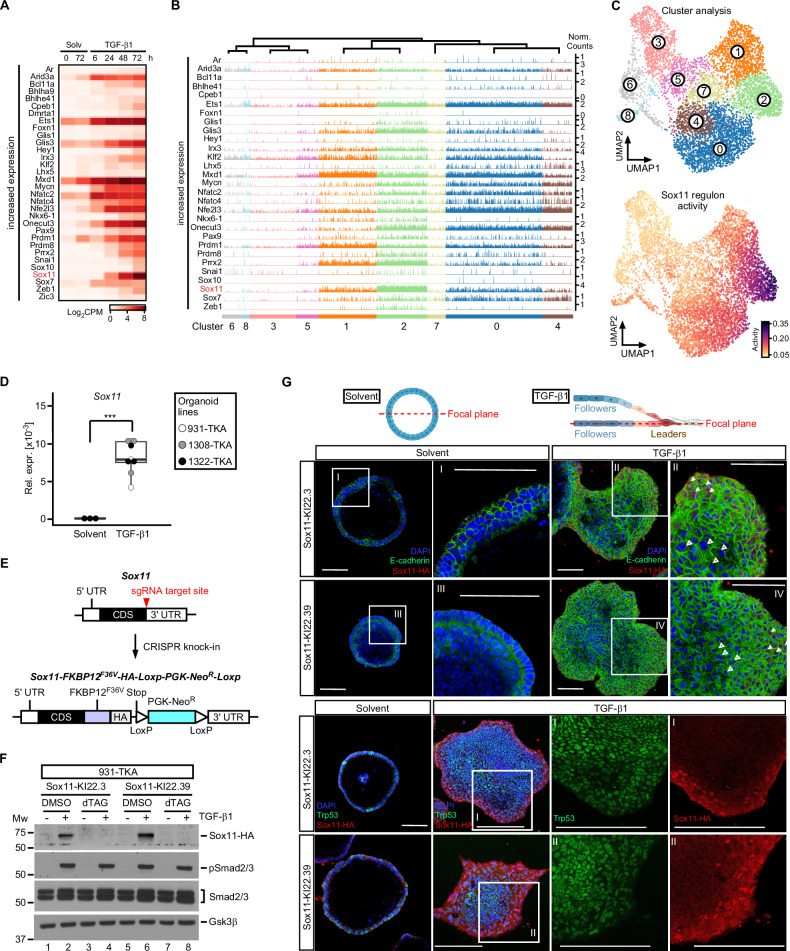
Fig. 3. Sox11 is the most highly expressed TGF-β1-inducible transcription factor in cluster 2 cells and localizes to a layer of cells at the invasive front of TKA-organoids.
A Heatmap displaying TF genes with increasing expression in combined, time-resolved bulk RNA-seq data from solvent (Solv) or TGF-β1-treated 931-TKA and 947-TKA organoids [10]. All murine TFs with log2 fold changes > 4 at least at one time point of TGF-β1 stimulation were selected for display. Samples treated with TGF-β1 for 6 h and 24 h were compared to samples harvested at the beginning of the treatment (solvent 0 h). Samples treated with TGF-β1 for 48 h and 72 h were compared to samples kept in solvent for 72 h. Normalized log2CPM values were used as gene expression units. B Trackplots showing cluster-wise expression of the same TF genes as in (A) in scRNA-seq data from solvent or TGF-β1-treated 931-TKA organoid cells. Each peak represents the normalized count for a given gene in a single organoid cell. C Unsupervised cell clustering and UMAP visualization of 931-TKA organoid cells upon TGF-β1 treatment for 0, 24, 48, and 72 h (top) and projection of SCENIC-predicted Sox11 regulon activity onto the UMAP (bottom). D Expression of Sox11 in TKA-organoid lines (931-TKA, 1308-TKA, and 1322-TKA). Organoids were embedded in 3 mg/ml Matrigel and treated with solvent or TGF-β1 for 72 h, followed by RNA collection and cDNA synthesis. RNA levels of Sox11 were evaluated by qRT-PCR and normalized by the expression levels of Eef1a1. The box plots show the 26th to 75th percentiles of the data and the median. Dots represent results of individual experiments (n = 3). One-Way ANOVA, ***: p value < 0.001. E Scheme depicting the strategy for CRISPR-mediated in-frame knock-in (KI) of FKBP12F36V coding sequences and the influenza virus hemagglutinin (HA)-epitope tag into the Sox11 gene. The upper and lower schemes represent WT and modified Sox11, respectively. The red arrowhead indicates the sgRNA target site. FKBP12F36V served as a degradation tag. The PGK-promoter driven neomycin resistance gene (NeoR) flanked by two loxP sites was used as selection marker. F Sox11 expression in Sox11-KI clones. Sox11-KI clones KI22.3 and KI22.39 were treated with solvent or TGF-β1 for 72 h. Two hours before harvest organoids additionally received DMSO or 500 nM dTAGV-1. Organoids were collected for western blot analysis to monitor expression of HA-tagged Sox11, phospho-Smad2/3 (pSmad2/3), and total Smad2/3. Gsk3β was detected to control equal loading. Representative results from one out of three experiments are shown (n = 3). MW: molecular weight standard in kDa. G 931-TKA Sox11-KI organoids were seeded in 3 mg/ml Matrigel and treated with solvent or TGF-β1 for 72 h, followed by whole-mount immunofluorescence staining of the antigens indicated and confocal microscopy. Nuclei were counterstained with DAPI. Positions of focal planes examined are shown in the schemes above the micrographs. Boxed areas with Roman numerals are shown at higher magnification. White arrowheads indicate nuclear Sox11-HA staining at organoid invasive fronts; empty arrowheads indicate absence of Sox11-HA staining in the organoid center. Scale bars: 100 µm. Representative images from one out of three experiments are shown (n = 3). The schemes were generated with BioRender.com.