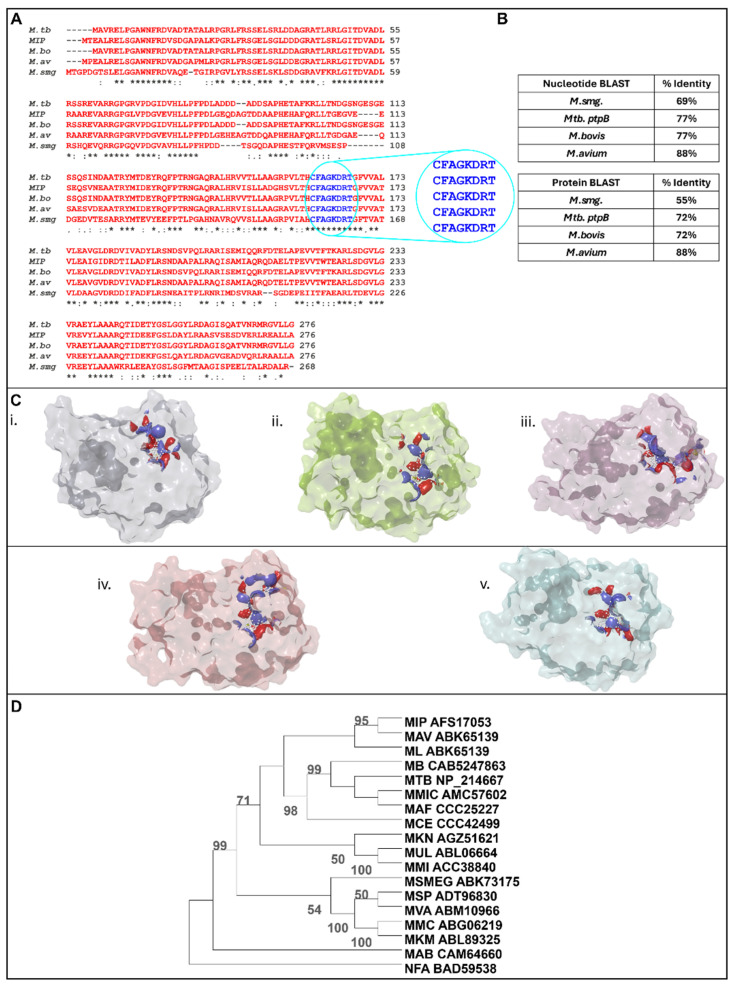
Figure 1.
The sequence of MIPPtpB (MIP_07528) is conserved among different mycobacterium species. (A) Alignment of protein tyrosine phosphatase B sequences of Mycobacterium tuberculosis H37Rv (NP_214667), Mycobacterium avium (WP_009979776), Mycobacterium bovis (CAB5247863) and Mycobacterium smegmatis (ABK73175) with Mycobacterium indicus pranii (AFS17053); the encircled region shows the active site signature. (* shows conserved base pairs and circle shows the variable region in sequence allignment) (B) Nucleotide and protein BLAST study among the identified protein tyrosine phosphatase B of Mtb, M. bovis, M. avium, M. smeg and the putative MIP_07528 to check the percentage identity among the pathogenic and the non-pathogenic mycobacterial species. (C) Active sites determined for (i) Rv0153c, (ii) MIP_07528, (iii) Mb0158c, (iv) MAV_5145 and (v) Msmeg_0100. (D) Phylogenetic tree based on the maximum likelihood statistical method using the bootstrap method phylogeny test, showing conservation of PtpB across the genus of Mycobacterium. The nodes that were supported by bootstrap values > 50% (1000 replicates) are shown. Nocardia farcinica (NFA) was used as the outgroup. Abbreviations—MIP, Mycobacterium indicus pranii; MAV, Mycobacterium avium complex; ML, Mycobacterium leprae; MB, Mycobacterium bovis; MTB, Mycobacterium tuberculosis H37Rv; MMIC, Mycobacterium tuberculosis variant microti; MAF, M. africanum; MCE, Mycobacterium canetti; MKN, Mycobacterium kansasii; MUL, Mycobacterium ulcerans; MMI, Mycobacterium marinum M; MSMEG, Mycobacterium smegmatis; MSP, Mycobacterium gilvum Spyr; MVA, Mycobacterium vanbaalenii PYR-1; MMC, Mycobacterium sp. MCS; MKM, Mycobacterium sp. KMS; MAB, Mycobacterium abscessus.