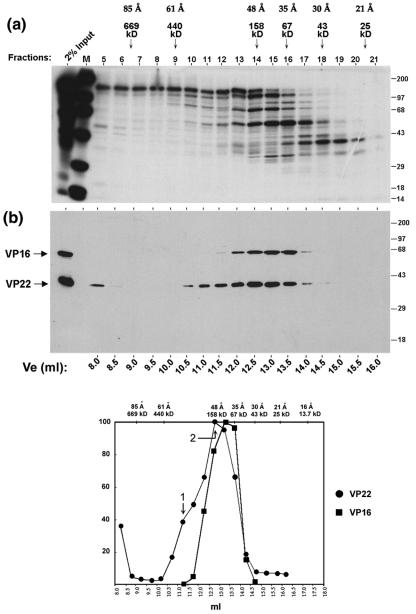
FIG. 3.
VP22 forms high molecular complexes in infected cells. (a) Autoradiograph of the elution profile from size exclusion chromatography of infected-cell extracts. High-salt extract prepared from HeLa cells infected with 5 PFU/cell of HSV-1 and labeled with 20 μCi of [35S]methionine from 12 to 16 h postinfection was fractionated by size exclusion chromatography as described in Materials and Methods using G-150 (150 mM salt buffer). One-tenth (50 μl) of each fraction (fractions 5 to 21) was separated in an SDS-PAGE gel and subjected to autoradiography. Lane M shows 14C-labeled protein standards and their positions are shown to the right of each panel. The first lane shows 2% of the input extract used in the separation analysis. The peak elution positions of the protein standards used to calibrate the size exclusion column are shown on the top of the panel along with their Stokes radii. (b) Western blot analysis of the fractions using AGV30 and LP1. A duplicate gel to that shown in panel a was electroblotted onto nitrocellulose and probed with anti-VP22 (AGV30) or anti-VP16 (LP1) antibodies. The elution volume of each fraction is shown at the bottom of the panel. The positions of VP16 and VP22 are shown to the left of the panel. (c) The elution profiles of VP22 (solid circles) and VP16 (solid squares) were measured by PhosphoImager densitometry analysis of the scanned blot and plotted as a percentage of the maximal peak versus elution volume. Peaks are numbered as discussed in the text. The elution positions of protein standards used in size exclusion chromatography are shown on the top of the graph along with their Stokes radii.