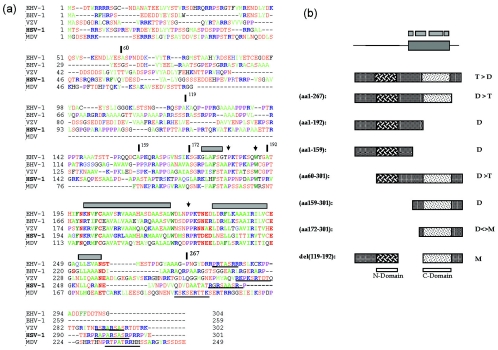
FIG. 8.
Conservation of a core domain and summary of VP22 oligomerization studies. (a) Sequences of VP22 homologues from equine herpesvirus type 1 (EHV-1), bovine herpesvirus type 1 (BHV-1), varicella-zoster virus (VZV), HSV-1, and Marek's disease virus (MDV) were aligned using the Clustal multiple-sequence alignment algorithm. The alignment was imported into the Genedoc program and alignments were optimized using information from all pairwise alignments. For ease of interpretation, residues have been color coded: A, F, I, L, M, V, W, and Y indicated in green; D, E, N, S, and T are in red; R and K are in blue; and P is in purple. Numbers relating to the boundaries of several HSV constructs used in this work are indicated with vertical bars above the sequence, while regions predicted to adopt secondary structure are indicated by gray boxes. A potential conserved motif towards the C-terminal end is underlined. (b) Summary of the oligomerization state of VP22 and various subregions. The top schematic indicate the conserved core with regions predicted from computer algorithms to adopt potential alpha-helical structure shown above. The preponderance of tetramers (T) and dimers (D) and monomers (M) is indicated for each VP22 species. Shading in two regions indicates the conclusion that VP22 contains two dimerization interfaces (N) and (C) as discussed in the text.