Abstract
The bovine herpesvirus 1 (BHV-1) tegument protein VP22 is predominantly localized in the nucleus after viral infection. To analyze subcellular localization in the absence of other viral proteins, a plasmid expressing BHV-1 VP22 fused to enhanced yellow fluorescent protein (EYFP) was constructed. The transient expression of VP22 fused to EYFP in COS-7 cells confirmed the predominant nuclear localization of VP22. Analysis of the amino acid sequence of VP22 revealed that it does not have a classical nuclear localization signal (NLS). However, by constructing a series of deletion derivatives, we mapped the nuclear targeting domain of BHV-1 VP22 to amino acids (aa) 121 to 139. Furthermore, a 4-aa motif, 130PRPR133, was able to direct EYFP and an EYFP dimer (dEYFP) or trimer (tEYFP) predominantly into the nucleus, whereas a deletion or mutation of this arginine-rich motif abrogated the nuclear localization property of VP22. Thus, 130PRPR133 is a functional nonclassical NLS. Since we observed that the C-terminal 68 aa of VP22 mediated the cytoplasmic localization of EYFP, an analysis was performed on these C-terminal amino acid sequences, and a leucine-rich motif, 204LDRMLKSAAIRIL216, was detected. Replacement of the leucines in this putative nuclear export signal (NES) with neutral amino acids resulted in an exclusive nuclear localization of VP22. Furthermore, this motif was able to localize EYFP and dEYFP in the cytoplasm, and the nuclear export function of this NES could be blocked by leptomycin B. This demonstrates that this leucine-rich motif is a functional NES. These data represent the first identification of a functional NLS and NES in a herpesvirus VP22 homologue.
Bovine herpesvirus 1 (BHV-1) is composed of four concentric compartments, the nucleoprotein core surrounding the double-stranded DNA, the capsid, the tegument, and the envelope (18). The tegument of alpha herpesviruses is defined as the amorphous region located between the virion capsid and the envelope and contains at least 15 viral gene products (20, 31). Not only are tegument proteins important viral structural proteins, but they also may play significant roles at several stages during virus infection. They are the first to interact with the intracellular environment and could exert their functions prior to viral gene expression and thus subjugate the host cell (22). At a later stage, the tegument proteins must associate to create the tegument of new virions (20).
BHV-1 VP22 is a 258-amino-acid (aa) tegument protein encoded by UL49 which has been shown to be dispensable for BHV-1 replication (16), although a BHV-1 VP22 deletion mutant yielded a lower titer than the wild-type virus in cell culture. Interestingly, this VP22 deletion mutant was asymptomatic and avirulent in cattle (15). Thus, VP22 might play an important role during BHV-1 infection. Previous studies have indicated that BHV-1 VP22 is predominantly localized in the nuclei of BHV-1-infected cells (16), which suggests that VP22 may have regulatory functions (16). However, the exact biological function of VP22 in infection is still unknown.
Similar to herpes simplex virus type 1 (HSV-1) VP22 (6), BHV-1 VP22 has the capability of intercellular spread, whereby the protein exits expressing cells and enters neighboring cells, where it is targeted to the nucleus (9, 32). BHV-1 VP22 interacts with histones and nucleosomes (28) and with microtubules and chromatin (9), and the carboxyl terminus (aa 118 to 258) of VP22 is required for nuclear localization (28).
The cell transports a variety of molecules into the nucleus, including metabolites and proteins as well as ribosomal subunits, certain RNAs, and ribonucleoproteins (8, 17, 24). The process of signal-mediated nuclear import is now well established for proteins. A typical nuclear protein contains a transferable basic nuclear localization signal (NLS), which is recognized by an importin (karyopherin) receptor (8). The karyopherin superfamily also includes a number of exportins responsible for nuclear export, which recognize a hydrophobic nuclear export signal (NES) (8). Small proteins of <40 to 60 kDa can passively diffuse through the nuclear pore complex, whereas larger proteins cannot move freely from the cytoplasm to the nucleus (8). The addition of an NLS to a small protein that passively diffuses through the nuclear pore complex or even to a larger protein confers to that protein all of the properties of NLS-dependent transport (11). In this study, we identify a functional NLS and NES in BHV-1 VP22.
MATERIALS AND METHODS
Cells and virus.
Madin-Darby bovine kidney (MDBK) cells were cultured in minimal essential medium (MEM; Gibco-BRL, Burlington, Ontario, Canada) supplemented with 10% fetal bovine serum (FBS; Gibco-BRL). COS-7 cells were grown in Dulbecco's modified MEM (DMEM; Gibco-BRL) supplemented with 10% FBS. The BHV-1 strain P8-2 (2) was used for the infection of MDBK cells and for viral DNA purification.
Construction of plasmids expressing VP22.
All enzymes used for cloning were purchased from Amersham Pharmacia Biotech (Baie d'Urfe, Quebec, Canada). The UL49 open reading frame was amplified from BHV-1 genomic DNA by a PCR using Deep Vent DNA polymerase (New England Biolabs Ltd., Mississauga, Ontario, Canada) and the primers 5′ GAGGAGAGATCTATGGCCCGGTTCCACAGGC 3′ and 5′ TAGAGGATCCGGCC GGGCCCGCTCGCCGCGAAG 3′. The product was digested with BglII and BamHI and inserted into the green fluorescent protein variant mammalian expression vector pEYFP-N1 (Clontech, BD Biosciences, Palo Alto, CA) encoding enhanced yellow fluorescent protein (EYFP), also cut with BglII and BamHI, to create pVP22-EYFP. The BHV-1 VP22 full-length coding region was cut from pVP22-EYFP with the restriction enzymes BglII and BamHI and subcloned into the green fluorescent protein variant mammalian expression vector pEYFP-C1 (Clontech) to create pEYFP-VP22. Subsequently, the VP22 full-length coding region, including the stop codon, was cut from pEYFP-VP22 with BglII and HincII, blunted by the Klenow fragment of DNA polymerase I (Amersham Pharmacia Biotech), and inserted into the eukaryotic expression vector pMASIA (12, 26), cut with EcoRI and blunted with the Klenow fragment of DNA polymerase I (Amersham Pharmacia Biotech), to create pMASIA-VP22.
Construction of plasmids encoding VP22 deletion derivatives.
A series of plasmids expressing VP22 with defined deletions were made as follows. Plasmid pVP22-EYFP was digested with PvuI and BglII and ligated with a linker comprised of the annealed oligonucleotides 5′ GATCTATGCGCGAT 3′ and 5′ CGCGCATA 3′ to generate a fragment consisting of aa 74 to 258 (F74-258). Plasmid pVP22-EYFP was digested with PpuMI and BglII and ligated with a linker comprised of the annealed nucleotides 5′ GATCTATGGG 3′ and 5′ GACCCCATA 3′ to generate F121-258. Plasmid pVP22-EYFP was digested with Van91I and BglII and ligated with a linker comprised of the annealed nucleotides 5′ GATCTATGGCCAGCGT 3′ and 5′ CTGGCCATA 3′ to generate F191-258. Plasmid pVP22-EYFP was digested with Van91I and AgeI and ligated with a linker comprised of the annealed oligonucleotides 5′ CTGGCCA 3′ and 5′ CCGGTG GCCAGACG 3′ to generate F1-190. Plasmid pVP22-EYFP was digested with PpuMI and AgeI, and the large fragment was ligated with a linker comprised of the annealed oligonucleotides 5′ GTCCTCCCCA 3′ and 5′ CCGGTGGGGAG 3′ to generate F1-120. To create F73-190, a DNA fragment (351 bp) cut from pVP22-EYFP with PvuI and Van91I was ligated with pEYFP-N1 digested with BglII and AgeI and with linkers comprised of the annealed nucleotides 5′ GATCTATGCGCGAT 3′ and 5′ CGCGCATA 3′ and the annealed nucleotides 5′ CTGGCCA 3′ and 5′ CCGGTGGCCAGACG 3′. To create F93-190, a DNA fragment (291 bp) cut from pVP22-EYFP with XhoI and Van91I was ligated with pEYFP-N1 digested with BglII and AgeI and with linkers comprised of the annealed nucleotides 5′ GATCTATGCGC 3′ and 5′ TCGAGCGCATA 3′ and the annealed nucleotides 5′ CTGGCCA 3′ and 5′ CCGGTGGCCAACG 3′. To create F140-190, a DNA fragment (153 bp) cut from pVP22-EYFP with FseI and Van91I was ligated with pEYFP-N1 digested with BglII and AgeI and with linkers comprised of the annealed nucleotides 5′ GATCTATGGGCCGG 3′ and 5′ CCCATA 3′ and the annealed nucleotides 5′ CTGGCCA 3′ and 5′ CCGGTGGCCAGACG 3′.
Construction of plasmids encoding VP22 NLS and NES deletion derivatives and domains.
Plasmids encoding aa 121 to 139 fused with EYFP (aa121 to 139-EYFP) or 130PRPR133 fused with EYFP (NLS-EYFP) were constructed by ligating the following annealed oligonucleotides with pEYFP-N1 (Clontech) cut with EcoRI and BamHI: 5′ AATTCATGGGCGCCGGGGCAGTCGCCGTCGGTCCACCTCGACCTCGCGCGCCCCCCGGTGCTAATCGG 3′ and 5′ GATCCCGATTAGCACCGGGGGGCGCGCGAGGTCGAGGTGGACCGACGGCGACTGCCCCGGCGCCCATG 3′ or 3′ AATTCATGCCTCGACCTCGG 5′ and 3′ GATCCCGAGGTCGAGGCATG 5′. A plasmid encoding VP22-EYFP with the putative NLS 130PRPR133 mutated to the neutral aa 130AAAA133 (VP22-mNLS-EYFP) was constructed from pVP22-EYFP by a PCR using the primers 5′ CGGAATTCATGGCCCGGTTCCACAGGCC 3′, 5′ TGGACCGACGGCGACTGCCC 3′, 5′ GCGCCCCCCGGTGCTAATGC 3′, and 5′ TGGCGACCGGTGGATCCGGC 3′. A plasmid encoding VP22-EYFP with the putative NLS 130PRPR133 deletion (VP22-dNLS-EYFP) was constructed from pVP22-EYFP by a PCR using the primers 5′ CGGAATTCATGGCCCGGTTCCACAGGCC 3′, 5′ TGCTGCTGCTGCTGGACCGACGGCGACTGCCC 3′, 5′ GCGCCCCCCGGTGCTAATGC 3′, and 5′ TGGCGACCGGTGG ATCCGGC 3′. A plasmid encoding an EYFP dimer (dEYFP) was constructed from pEYFP-N1 by a PCR using the following primers: 5′ CGGAATTCATGGTGAGCAAGGGCGAGGA 3′ and 5′ GAAGATCTTTGTACAGCTCGTCCATGC 3′. The PCR product was digested with EcoRI and BamHI and inserted into pEYFP-N1 cut with the same enzymes to create pdEYFP. A plasmid encoding the NLS fused with dEYFP (NLS-dEYFP) was constructed from pNLS-EYFP by a PCR using the primers 5′ CAAGCTTCGAATTCATGCCT 3′ and 5′ GAAGATCTTTGTACAGCTCGTCCATGC 3′. The PCR product was digested with EcoRI and BamHI and inserted into pEYFP-N1 cut with the same enzymes to create pNLS-dEYFP. A plasmid encoding an EYFP trimer (tEYFP) or the NLS fused with tEYFP (NLS-tEYFP) was constructed similarly.
A plasmid encoding VP22-EYFP with the putative NES 204LDRMLKSAAIRIL216 mutated to the neutral aa 204ADRMAKSAAARAA216 (VP22-mNES-EYFP) was constructed from pVP22-EYFP by a PCR using the primers 5′ CGGAATTCATGGCCCGGTTCCACAGGCC 3′ and 5′ TCGCTCGTTGCTCTTTGGGG 3′ or 5′ GCAGATCGCATGGCAAAGTCGGCGGCAGCACGCGCAGCAGTGTGCGAGGGCTCCGGG 3′ and 5′ TGGCGACCGGTGGATCCGGC 3′. A plasmid encoding the putative NES fused with EYFP (NES-EYFP) was constructed by ligating the following annealed oligonucleotides with pEYFP-N1 (Clontech) cut with EcoRI and BamHI: 5′ AATTCATGTTGGATCGCATGTTGAAGTCGGCGGCAATTCGCATCCTCGTG 3′ and 5′ GATCCACGAGGATGCGAATTGCCGCCGACTTCAACATGCGATCCAACATG 3′. A plasmid encoding the NES fused with dEYFP (NES-dEYFP) was constructed from pNES-EYFP by a PCR with the primers 5′ CAAGCTTCGAATTCATGTTG 3′ and 5′ GAAGATCTTTGTACAGCTCGTCCATGC 3′. The PCR product was digested with EcoRI and BamHI and inserted into pEYFP-N1 cut with the same enzymes to create pNES-dEYFP. All constructs were verified by sequencing.
Transfection.
To express the proteins in vitro, COS-7 cells were plated onto six-well plates (Corning, Corning, NY) in DMEM (Gibco-BRL) with 10% FBS at a density of 2.5 × 105 cells per well overnight before transfection. Transfection mixtures, consisting of 1.0 to 1.5 μg of plasmid and Lipofectamine Plus reagent (Gibco-BRL), were prepared according to the manufacturer's instructions. Briefly, the plasmids were diluted in Optimem (Gibco-BRL) containing Lipofectamine Plus and incubated for 15 min at room temperature. Optimen containing Lipofectamine was added to the mixture and further incubated for 15 min at room temperature (RT). COS-7 cells were washed with Optimem, and the transfection mixture, made up to 1 ml with Optimem, was added to the cells and incubated at 37°C in a humidified 5% CO2 incubator for 5 h, after which an equal volume of DMEM containing 10% FBS was added. Leptomycin B (LMB; Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada) was used in some experiments at 20 ng ml−1 after transfection.
Cell fractionation.
The nuclear and cytoplasmic fractions were isolated as described previously (29, 30). In brief, transfected COS-7 cells were lysed with NP-40-containing lysis buffer (10 mM Tris, pH 7.4, 10 mM NaCl, 5 mM MgCl2, 0.5% NP-40) to disrupt the cell membrane, and the cell lysate was centrifuged at 500 × g for 5 min at 4°C. The supernatant containing the cytoplasmic fraction was removed, and the pellet containing the nuclear fraction was washed gently in 1 ml of lysis buffer to remove any remaining cytoplasmic material and then pelleted again. Both fractions were subjected to Western blot analysis.
Western blot analysis.
To confirm protein expression, the plasmid-transfected COS-7 cells were collected in cell lysis buffer (0.05 M Tris, pH 8.0, 0.15 M NaCl, 0.1% sodium dodecyl sulfate [SDS], 1% NP-40, 1% deoxycholine) 24 h after transfection. Equivalent amounts of each cell lysate were separated by SDS-polyacrylamide gel electrophoresis on an 8.5% gel, transferred to a nitrocellulose membrane, and incubated with EYFP-specific polyclonal rabbit antibody (Clontech) and/or VP22-specific polyclonal rabbit antibody. Alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Kirkegaard and Perry Laboratories, Gaithersburg, Maryland) was used as the secondary antibody. Reactive bands were revealed with nitro blue tetrazolium bromochlorindolyl phosphate tablets (Sigma-Aldrich). The Western blots were scanned and processed using Adobe Photoshop.
Fluorescence microscopy.
For immunofluorescence, cells were either observed live or fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS; 0.137 M NaCl, 0.003 M KCl, 0.008 M Na2HPO4, 0.001 M NaH2PO4, pH 7.4) for 20 min, washed three times with PBS, and permeabilized with 0.5% Triton X-100 in PBS for 10 min. The cells were rinsed with PBS and then incubated with PBS containing 10% FBS for 20 min at RT. Subsequently, VP22-specific polyclonal antibody (16) diluted in PBS containing 10% FBS was added to the cells, which were again incubated for 20 min at RT. Finally, fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (Zymed Laboratories, San Francisco, CA) in PBS containing 10% FBS was added, followed by a 20-min incubation at RT. After each incubation step, the cells were washed extensively with PBS. The cells were mounted in Vectashield (Vector Laboratories, Burlingame, CA). Fixed stained cells and live cells were analyzed using a Zeiss Axiovert 200 M microscope (Carl Zeiss Canada Ltd., Toronto, Ontario, Canada). The subcellular localization patterns in the live cells transfected with VP22-EYFP or VP22 deletion mutants linked to EYFP were quantified by counting 100 cells; the predominant pattern was described and reported as a percentage of the total number of cells counted. The images were captured and processed using Adobe Photoshop.
RESULTS
Subcellular localization of VP22 and VP22-EYFP.
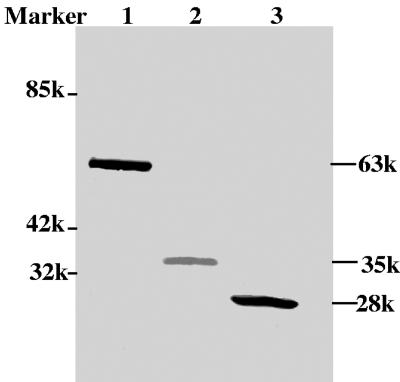
Liang and colleagues have demonstrated that VP22 is predominantly localized in the nuclei of BHV-1-infected cells (16). To confirm the subcellular localization of VP22 in the absence of other viral proteins, the UL49 gene was cloned into the eukaryotic expression vectors pMASIA (12, 26) and pEYFP-N1, such that the UL49 gene is fused in-frame to the N terminus of the EYFP gene. Western blotting of COS-7 cells transfected with a plasmid encoding VP22-EYFP, EYFP, or VP22 demonstrated that BHV-1 VP22 had an apparent molecular mass of 35 kDa, as expected (16), whereas VP22-EYFP had an apparent molecular mass of 63 kDa, which corresponds to the combined apparent molecular masses of VP22 and EYFP (Fig. 1).
FIG. 1.
Analysis of VP22-EYFP, VP22, and EYFP in transiently transfected COS-7 cells by Western blotting. The proteins were probed with both VP22- and EYFP-specific polyclonal antibodies. The molecular weight of each protein is indicated in the right margin. Lane 1, VP22-EYFP; lane 2, VP22; lane 3, EYFP. Mr standards are shown in the left margin.
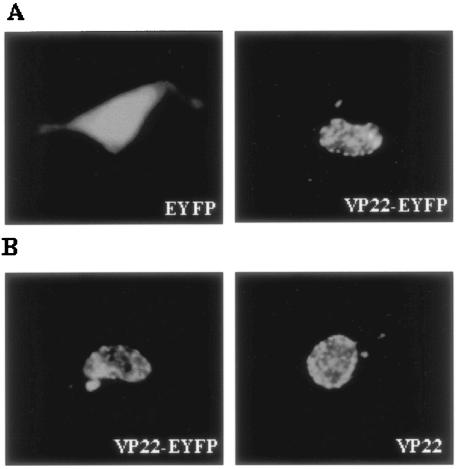
To study the cellular localization of VP22 in the absence of other viral proteins, COS-7 cells were transfected with a plasmid encoding EYFP, VP22-EYFP, or VP22 and examined live by fluorescence microscopy (Fig. 2A) or after fixation by immunofluorescence using VP22-specific antibodies (16) (Fig. 2B). Representative images of cells transfected with plasmids encoding VP22-EYFP, EYFP, and VP22 are shown. As expected, diffuse fluorescence was observed throughout both the cytoplasm and the nucleus in the pEYFP-N1-transfected cells at all times (Fig. 2A). In contrast, fluorescence in pVP22-EYFP-transfected cells was localized predominantly within the nucleus (Fig. 2A), with small amounts of VP22-EYFP present in the cytoplasm. This was typical for 88% of the observed cells and is similar to the results reported previously (9, 28). In cells that showed VP22-EYFP in the cytoplasm, filamentous labeling was sometimes observed, as described previously (9). The subcellular localization of VP22 in fixed cells transfected with pMASIA-VP22, a eukaryotic vector expressing VP22, appeared to be similar to that observed in live cells transfected with pVP22-EYFP (Fig. 2B), which confirmed that VP22-EYFP behaves in an identical manner to that of untagged VP22, thus validating VP22-EYFP as a suitable system for investigating the NLS and NES of VP22.
FIG. 2.
Subcellular localization of VP22 in transfected cells. (A) Subcellular distribution of EYFP and VP22-EYFP. COS-7 cells were transiently transfected with pEYFP-N1 or pVP22-EYFP, and live cells were examined by fluorescence microscopy 24 h after transfection. (B) Subcellular distribution of VP22-EYFP and VP22. COS-7 cells were transiently transfected with pVP22-EYFP or pMASIA-VP22, a eukaryotic vector expressing VP22, fixed, and incubated with VP22-specific polyclonal antibodies to identify VP22. Images of representative cell fields are shown. Magnification, ×40.
Subcellular localization of VP22 deletion constructs.
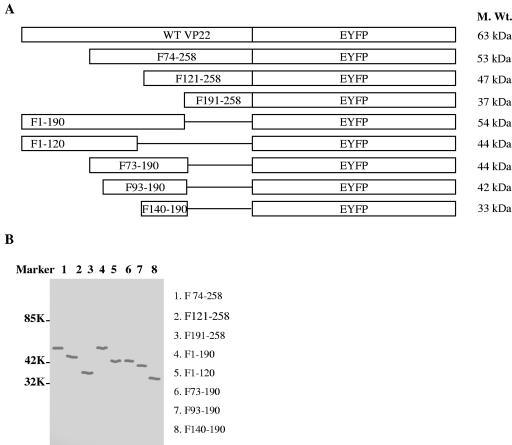
To identify the domains responsible for the nuclear localization of VP22, deletion derivatives of VP22-EYFP were constructed as depicted in Fig. 3A. To confirm the correct expression of the deletion mutants of pVP22-EYFP, Western blotting of the transfected COS-7 cell lysates was performed using antibodies to VP22 (Fig. 3B). All mutants expressed proteins of the predicted molecular weights. COS-7 cells were transfected with these constructs, and the distribution of the VP22-EYFP mutant proteins was analyzed 24 h and 48 h after transfection in live cells by fluorescence microscopy. The cellular localization patterns of live cells at 24 h and 48 h were found to be identical, with the exception of the intercellular trafficking property, which was not detected until 48 h after transfection, as described previously (32). The deletion mutant constructs each had a distinct localization pattern. Figure 4 shows typical images of cells that are representative of each mutant at 24 h.
FIG. 3.
Construction and analysis of expression of deletion derivatives of VP22. (A) Schematic diagram of full-length VP22-EYFP and the truncated mutants used for this study. (B) Deletion derivatives of VP22 identified by Western blotting of transiently transfected COS-7 cells. The proteins were probed with VP22-specific polyclonal antibodies. The VP22 fragments are indicated in the right margin. Mr standards are shown in the left margin.
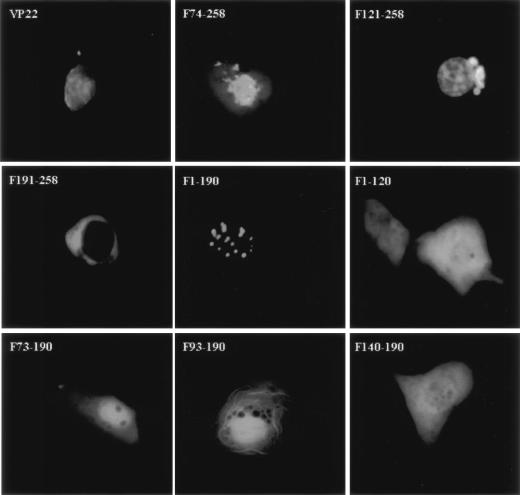
FIG. 4.
Subcellular localization of VP22-EYFP deletion derivatives in live cells. COS-7 cells were transiently transfected with the deletion constructs and observed live 24 h after transfection by fluorescence microscopy. Each image is representative of the majority of the cells observed in several fields. Magnification, ×40.
In COS-7 cells transfected with the plasmid encoding VP22 F74-258, fluorescence was primarily found in the nucleus, with some found in the cytoplasm. This was typical for 81% of the transfected cells. In 78% of plasmid-transfected cells, VP22 F121-258 was present predominantly in the nucleus, where it displayed a diffuse pattern; in addition, a perinuclear mass was observed. In contrast, VP22 F191-258 localized exclusively in the cytoplasm of 95% of the transfected COS-7 cells. In COS-7 cells transfected with the plasmid encoding VP22 F1-190, distinct fluorescent speckles were observed in the nucleus. This applied to 97% of the transfected cells. VP22 F1-120 was evenly distributed between the cytoplasm and the nucleus in 98% of the transfected cells, similar to EYFP alone. Although VP22 F73-190 was also found in both the cytoplasm and the nucleus, it showed a predominantly nuclear localization in 91% of the cells. Similarly, VP22 F93-190 was present in both the nucleus and the cytoplasm, but with a pronounced filamentous pattern which was likely due to binding to the cytoplasmic skeleton (9). This was observed for 83% of the counted cells. Finally, VP22 F140-190 was localized diffusely in the cytoplasm and the nucleus in 94% of the transfected cells.
Identification of an NLS responsible for nuclear localization.
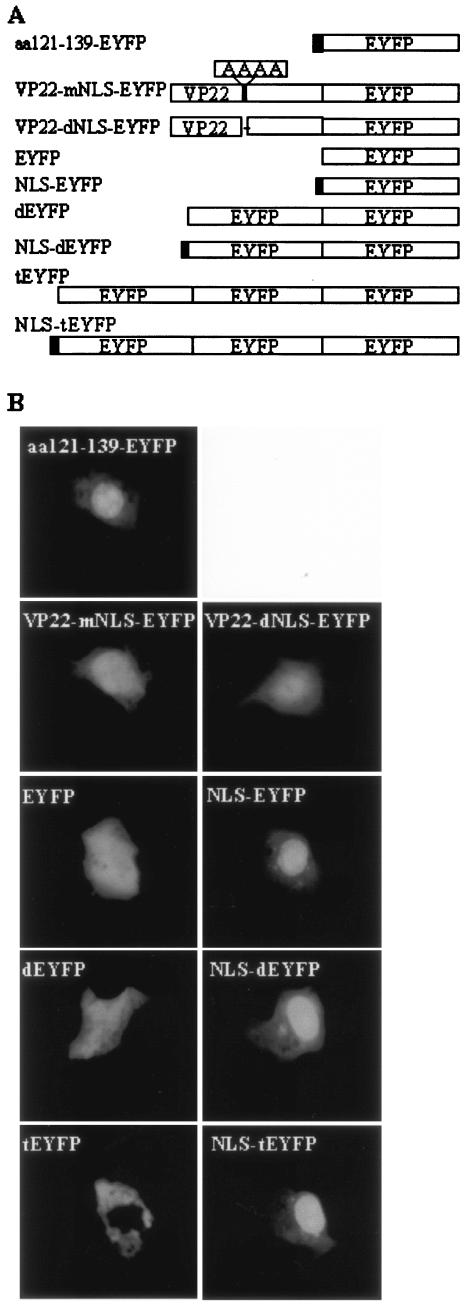
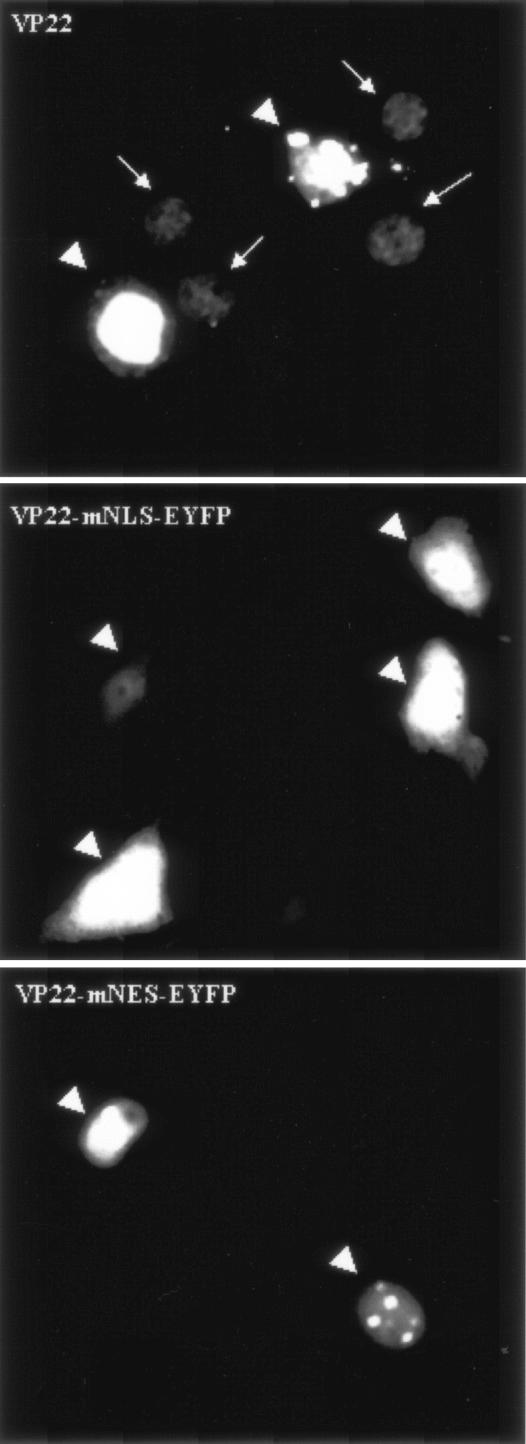
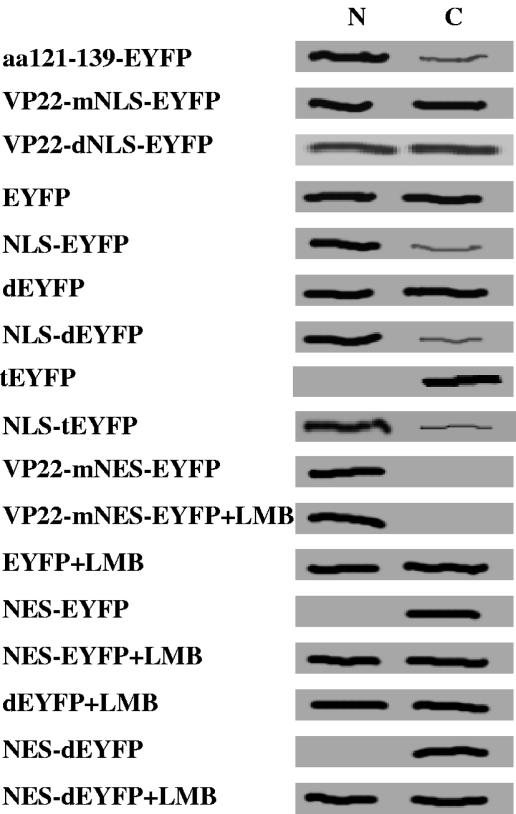
Additional studies are required to explain the distinct patterns observed for each mutant. Nuclear localization was observed for F74-258, F121-258, F1-190, F73-190, and F93-190, but not for F1-120, F140-190, and F191-258; therefore, amino acid residues 121GGAGAVAVGPPRPRAPPGA139 (aa 121 to 139) were considered to be essential for nuclear localization. Since VP22 is targeted to the nucleus (16, 28) and interacts with histones and nucleosomes (28), it has been suggested that the nuclear localization of VP22 is an important characteristic and that VP22 may have a modulatory function during BHV-1 infection. This prompted us to focus on further characterizing the putative nuclear localization domain of aa 121 to 139. To confirm that aa 121 to 139 mediate the nuclear localization of VP22, a plasmid encoding EYFP fused to aa 121 to 139 from VP22 was constructed (aa121 to 139-EYFP; Fig. 5A), and the subcellular localization of the aa121-139-EYFP fusion protein was examined 24 h after transfection in live COS-7 cells by fluorescence microscopy. As expected from the deletion mutant analysis, aa121-139-EYFP was directed predominantly, though not entirely, into the nucleus (Fig. 5B). An arginine-rich motif, 130PRPR133, which showed considerable similarity to the NLS (RRPR) of BHV-1 VP8 (33), was identified in the amino acid sequence of VP22. The replacement of 130PRPR133 with the neutral aa residues 130AAAA133 (VP22-mNLS-EYFP) or the deletion of 130PRPR133 (VP22-dNLS-EYFP) (Fig. 5A) abrogated the nuclear localization of VP22 and resulted in the same pattern of subcellular localization as that observed for the reporter protein EYFP alone (Fig. 5B). Furthermore, a construct was made which expressed EYFP N-terminally linked to the putative NLS, 130PRPR133 (NLS-EYFP; Fig. 5A). The NLS-EYFP fusion protein localized primarily to the nucleus, similar to aa121-139-EYFP (Fig. 5B) and the RRPR-EYFP fusion protein reported previously (33). To enhance the sensitivity of the test and to confirm nuclear transport for a larger protein that does not diffuse freely in the cell, NLS-dEYFP (∼56 kDa) and NLS-tEYFP (∼84 kDa), which express the putative VP22 NLS fused to an EYFP dimer (dEYFP) and trimer (tEYFP), respectively, were constructed (Fig. 5A). Although dEYFP was still small enough to be distributed throughout the nucleus and cytoplasm, tEYFP remained entirely cytoplasmic, demonstrating that it cannot move into the nucleus (Fig. 5B). The 130PRPR133 motif mediated the nuclear localization of both dEYFP and tEYFP, which further confirms the ability of the 130PRPR133 motif to mediate nuclear localization of a heterologous protein (Fig. 5B). Interestingly, the mutation of the NLS 130PRPR133 also eliminated the ability of VP22 to spread intercellularly (Fig. 6). An analysis by Western blotting of the nuclear and cytoplasmic fractions of COS-7 cells transfected with plasmids encoding EYFP, dEYFP, tEYFP, NLS-EYFP, NLS-dEYFP, and NLS-tEYFP further supported the results observed by fluorescence microscopy (Fig. 7). Collectively, these results indicate that the 130PRPR133 sequence of VP22 constitutes a functional NLS.
FIG. 5.
Identification of an NLS responsible for nuclear localization. (A) Schematic diagram of the EYFP fusion protein used to identify the VP22 NLS. The names of the constructs are shown on theleft. (B) COS-7 cells were transiently transfected with plasmids encoding aa121-139-EYFP, VP22-mNLS-EYFP, VP22-dNLS-EYFP, EYFP, NLS-EYFP, dEYFP, NLS-dEYFP, tEYFP, and NLS-tEYFP and examined live 24 h after transfection by fluorescence microscopy. Each image is representative of the majority of the cells observed in several fields. Magnification, ×40.
FIG. 6.
Effect of mutation of the NLS or NES on intracellular trafficking of VP22. COS-7 cells were transfected with plasmids encoding VP22-EYFP, VP22-mNLS-EYFP, and VP22-mNES-EYFP and examined live 48 h after transfection by fluorescence microscopy. Original VP22-expressing cells are indicated with arrowheads, and the neighboring cells containing VP22 are indicated with arrows. Each image is representative of the majority of the cells observed in several fields. Magnification, ×40.
FIG. 7.
Analysis of EYFP distribution in nuclear (N) and cytoplasmic (C) fractions of transfected COS-7 cells. COS-7 cells were transfected with plasmids encoding aa121-139-EYFP, VP22-mNLS-EYFP, VP22-dNLS-EYFP, EYFP, NLS-EYFP, dEYFP, NLS-dEYFP, tEYFP, NLS-tEYFP, VP22-mNES-EYFP, VP22-mNES-EYFP (plus LMB treatment), EYFP (plus LMB treatment), NES-EYFP, NES-EYFP (plus LMB treatment), dEYFP (plus LMB treatment), NES-dEYFP, and NES-dEYFP (plus LMB treatment). At 24 h posttransfection, COS-7 cells were fractionated into nuclear and cytoplasmic fractions as described previously (29, 30). Equivalent amounts of each fraction were separated by SDS-polyacrylamide gel electrophoresis on an 8.5% gel, transferred to nitrocellulose, and probed with an EYFP-specific polyclonal antibody.
Identification of an NES in VP22.
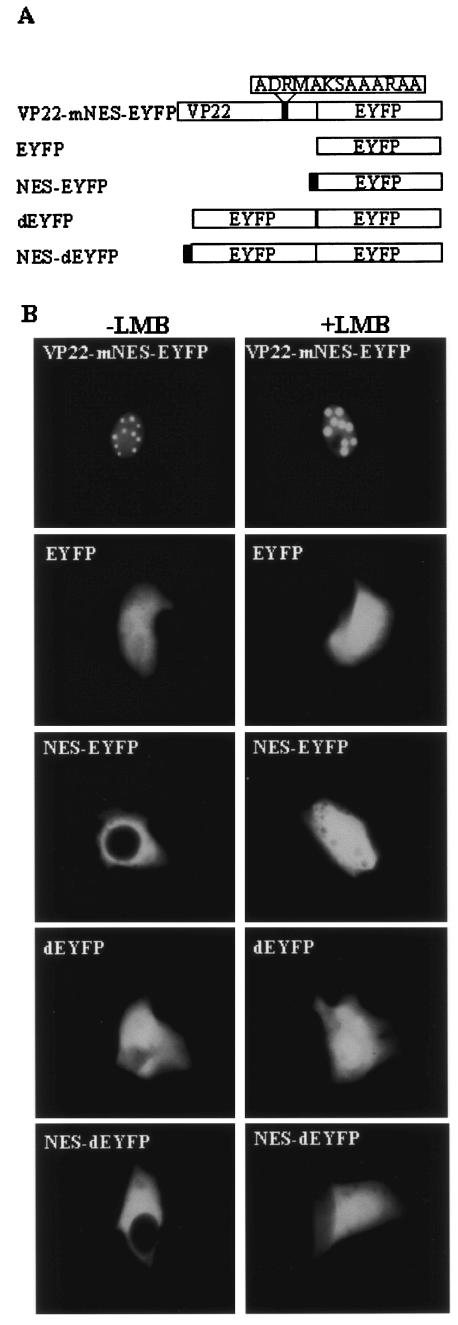
Nuclear export signals consist of largely hydrophobic, leucine-rich sequences (24). The cytoplasmic localization of VP22 F1-190 suggests that there might be a nuclear export domain in the C terminus (aa 191 to 258) of VP22. An examination of the C-terminal region of VP22 revealed that it contains a leucine-rich motif, 204LDRMLKSAAIRIL216, which was speculated to be a potential NES. To validate this hypothesis, the putative NES was mutated by replacing the leucine residues with neutral alanine residues to create 204ADRMAKSAAARAA216 (VP22-mNES-EYFP; Fig. 8A). COS-7 cells were transfected with VP22-mNES-EYFP and examined live by fluorescence microscopy. As anticipated, VP22-mNES-EYFP was exclusively localized in the nucleus (Fig. 8B), similar to the nuclear localization pattern observed for VP22 F1-190. To determine whether the NES alone can direct nuclear export, a plasmid was constructed which expressed the NES fused to the N termini of EYFP and dEYFP (NES-EYFP and NES-dEYFP; Fig. 8A). An examination by fluorescence microscopy of live COS-7 cells transfected with the plasmids encoding NES-EYFP and NES-dEYFP revealed that the NES localized all of the EYFP and dEYFP to the cytoplasm (Fig. 8B).
FIG. 8.
Identification of an NES in VP22. (A) Schematic diagram of the EYFP fusion proteins used to identify the VP22 NES. Thenames of the constructs are shown on the left. (B) COS-7 cells were transiently transfected with plasmids encoding VP22-mNES-EYFP, EYFP, NES-EYFP, dEYFP, and NES-dYEFP, with or without treatment with LMB, and examined live 24 h after transfection by fluorescence microscopy. Each image is representative of the majority of the cells observed in several fields. Magnification, ×40.
LMB, a potent and specific nuclear export inhibitor (7), alkylates and inhibits chromosome region maintenance 1 (CRM-1) or exportin 1, a protein required for the nuclear export of proteins containing an NES (13). Figure 8B shows that the nuclear export activity of the NES was completely abolished by a treatment with LMB at 20 ng ml−1 after transfection, thereby changing the subcellular localization of NES-EYFP or NES-dEYFP to an equal distribution in the nucleus and the cytoplasm. However, LMB did not affect the subcellular localization of EYFP, dEYFP, and VP22-mNES-EYFP, indicating that the export of NES-EYFP and NES-dEYFP from the nucleus to the cytosol is CRM-1 dependent. These observations were confirmed by Western blotting of the nuclear and cytoplasmic fractions of COS-7 cells transfected with a plasmid encoding dEYFP or dEYFP fused to the VP22 NES (Fig. 7). Interestingly, the NES mutation also resulted in abrogation of the intercellular trafficking property of VP22 (Fig. 6).
DISCUSSION
The subcellular localization of BHV-1 VP22 was investigated by the transient expression of VP22-EYFP. The transfection of COS-7 cells with pVP22-EYFP or pMASIA-VP22 demonstrated that VP22 localized predominantly to the nucleus in the absence of other viral proteins, which is consistent with observations with BHV-1-infected MDBK cells (16) and transfected D17 cells (9, 28).
The exact function of VP22 during BHV-1 infection remains unknown. It is likely that various dynamic interactions of VP22 with different cellular proteins are required. VP22 has been shown to associate with histones and nucleosomes and may have modulatory effects on virus replication (28); the nuclear localization of VP22 must play an important role in this context, and this prompted us to identify the NLS of VP22.
The nuclear localization of HSV-1 VP22 has been suggested to modulate the transcription or replication of host cells, perhaps preparing them for infection (5, 6). Sequence analysis using PSORT II (23; http://psort.nibb.ac.jp) predicts that VP22 does not have a classical NLS. However, there are discernible clusters of basic amino acids, which may serve as an NLS (27). Furthermore, according to the PSORT II's k-nearest-neighbor algorithm prediction (10), BHV-1 VP22 has a 73.9% reliability to target to the nucleus, whereas HSV-1 VP22 has only a 34.8% reliability to target to the nucleus. We constructed a series of deletion mutants of BHV-1 VP22 fused to the N terminus of EYFP. EYFP has previously been used to tag the VP22 proteins of BHV-1 and HSV-1 (5, 6, 9, 28, 32) and did not affect the subcellular localization of VP22 in this study (Fig. 2). The subcellular localization patterns of the VP22 deletion mutants generally resembled those previously reported for similar constructs (28). Indeed, F1-120 and VP22 (aa 1 to 123) (28) were evenly distributed throughout the nucleus and cytoplasm, whereas F121-258 and VP22 (aa 118 to 258) (28) were both localized in the nucleus. There was one difference, in that F1-190 was targeted to the nucleus with a speckled pattern in our study, whereas VP22 (aa 1 to 197) localized to the nucleus and the cytoplasm (28). Moreover, in this study we identified a functional NLS in BHV-1 VP22, namely, 130PRPR133. Thus, our results and those of others (9, 16, 28) suggest that VP22 has a stronger nuclear localization ability than that of HSV-1 VP22.
The observation that the C-terminal portion of VP22 localized exclusively to the cytoplasm prompted us to reexamine its amino acid sequence, and a leucine-rich region was found in the C terminus (204LDRMLKSAAIRIL216). In addition, we demonstrated this sequence to be a functional NES, and its nuclear export activity was completely blocked by LMB. Thus, VP22 is an LMB-sensitive protein, which suggested that VP22 mediates nuclear export through a leucine-rich NES by a CRM-1-dependent pathway. The mutants demonstrated that BHV-1 VP22 has an NLS and an NES, which suggests that VP22 is a virus-encoded nucleocytoplasmic shuttle protein, as are BHV-1 VP8 (33) and other viral proteins (1, 3, 4, 14, 19, 21). By alignment, the highest level of homology was found between the NES of VP22 and the Rex protein of human T-cell leukemia virus type I (LSAQLYSSLSLD) (1).
Interestingly, NLS mutation or deletion abrogated nuclear localization, but the fluorescence did not accumulate in the cytoplasm, as one might expect due to the presence of an NES. One possibility is that protein folding of wild-type VP22 or VP22 with an NLS mutation might conceal the ability of the NES to bind to CRM-1 and thus the function of the NES, whereas most of VP22 was removed in VP22 F191-258, thus possibly enhancing the function of the NES. A similar phenomenon has been observed for the BHV-1 tegument protein VP8 (33).
BHV-1 VP22 has a marked capacity for intercellular trafficking, and it can improve the immune response to a DNA vaccine both in mice (25) and in cattle (32). Because of its potential importance both in virus infection and as a therapeutic transporter, we investigated the existence of nuclear transport sequences in VP22 and identified a functional NLS and NES of VP22. Furthermore, we demonstrated that mutation of either region resulted in the loss of the intercellular trafficking property of VP22 (Fig. 6), suggesting that both are essential for this property of BHV-1 VP22. It has been reported that HSV-1 VP22 binds to a cell surface receptor linked to actin microfilaments and enters cells via a novel pathway involving the cytoskeleton; after entering cells, VP22 is imported into the nucleus, despite lacking a nuclear localization signal (6). However, a putative NLS, 295RPRR298, exists in the C terminus of HSV-1 VP22, as predicted by PSORT II (23). Since the C-terminal 34 aa residues have been shown to be essential for the intercellular trafficking property of HSV-1 VP22 (6), the 295RPRR298 sequence may also be a crucial motif required for the intercellular spreading property of HSV-1 VP22, given that the 130PRPR133 motif of BHV-1 VP22 was demonstrated in this study to be required for this property. In conclusion, we have identified an NLS and an NES in BHV-1 VP22, and our results demonstrated that both are important for trafficking of VP22.
Acknowledgments
We thank Brenda Karvonen, Laura Latimer, and Marlene Snider for technical assistance.
This work was funded by the Natural Science and Engineering Research Council, Canadian Institutes of Health Research, Alberta Agricultural Research Institute, and Agricultural Development Fund of Saskatchewan. L. A. Babiuk is a recipient of a Canada research chair in vaccinology. A Saskatchewan Health Research Foundation fellowship was awarded to C. Zheng.
Footnotes
Published as VIDO's Journal Series number 384.
REFERENCES
- 1.Bogerd, H. P., R. A. Fridell, R. E. Benson, J. Hua, and B. R. Cullen. 1996. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 16:4207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter, D. E., and V. Misra. 1991. The most abundant protein in bovine herpes 1 virions is a homologue of herpes simplex virus type 1 UL47. J. Gen. Virol. 72:3077-3084. [DOI] [PubMed] [Google Scholar]
- 3.Chen, L., G. Liao, M. Fujimuro, O. J. Semmes, and S. D. Hayward. 2001. Properties of two EBV Mta nuclear export signal sequences. Virology 288:119-128. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, G., M. E. Brett, and B. He. 2002. Signals that dictate nuclear, nucleolar, and cytoplasmic shuttling of the gamma(1)34.5 protein of herpes simplex virus type 1. J. Virol. 76:9434-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott, G., and P. O'Hare. 2000. Cytoplasm-to-nucleus translocation of a herpesvirus tegument protein during cell division. J. Virol. 74:2131-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308-311. [DOI] [PubMed] [Google Scholar]
- 8.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 9.Harms, J. S., X. Ren, S. C. Oliveira, and G. A. Splitter. 2000. Distinctions between bovine herpesvirus 1 and herpes simplex virus type 1 VP22 tegument protein subcellular associations. J. Virol. 74:3301-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton, P., and K. Nakai. 1997. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5:147-152. [PubMed] [Google Scholar]
- 11.Jans, D. A., and S. Hubner. 1996. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol. Rev. 76:651-685. [DOI] [PubMed] [Google Scholar]
- 12.Krieg, A. M., T. Wu, R. Weeratna, S. M. Efler, L. Love-Homan, L. Yang, A. K. Yi, D. Short, and H. L. Davis. 1998. Sequence motifs in adenoviral DNA block immune activation by stimulatory CpG motifs. Proc. Natl. Acad. Sci. USA 95:12631-12636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, Y., Y. Yamakita, and R. M. Krug. 1998. Regulation of a nuclear export signal by an adjacent inhibitory sequence: the effector domain of the influenza virus NS1 protein. Proc. Natl. Acad. Sci. USA 95:4864-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang, X., B. Chow, and L. A. Babiuk. 1997. Study of immunogenicity and virulence of bovine herpesvirus 1 mutants deficient in the UL49 homolog, UL49.5 homolog and dUTPase genes in cattle. Vaccine 15:1057-1064. [DOI] [PubMed] [Google Scholar]
- 16.Liang, X., B. Chow, Y. Li, C. Raggo, D. Yoo, S. Attah-Poku, and L. A. Babiuk. 1995. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL49 homolog is dispensable for virus growth. J. Virol. 69:3863-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol Rev. 65:570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayfield, J. E., P. J. Good, H. J. VanOort, A. R. Campbell, and D. E. Reed. 1983. Cloning and cleavage site mapping of DNA from bovine herpesvirus 1 (Cooper strain). J. Virol. 47:259-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mears, W. E., and S. A. Rice. 1998. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology 242:128-137. [DOI] [PubMed] [Google Scholar]
- 20.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer, B. E., J. L. Meinkoth, and M. H. Malim. 1996. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: identification of a family of transferable nuclear export signals. J. Virol. 70:2350-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrison, E. E., A. J. Stevenson, Y. F. Wang, and D. M. Meredith. 1998. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J. Gen. Virol. 79:2517-2528. [DOI] [PubMed] [Google Scholar]
- 23.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-36. [DOI] [PubMed] [Google Scholar]
- 24.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira, S. C., J. S. Harms, R. R. Afonso, and G. A. Splitter. 2001. A genetic immunization adjuvant system based on BVP22-antigen fusion. Hum. Gene Ther. 12:1353-1359. [DOI] [PubMed] [Google Scholar]
- 26.Pontarollo, R. A., L. A. Babiuk, R. Hecker, and S. van Drunen Littel-van den Hurk. 2002. Augmentation of cellular immune responses to bovine herpesvirus-1 glycoprotein D by vaccination with CpG-enhanced plasmid vectors. J. Gen. Virol. 83:2973-2981. [DOI] [PubMed] [Google Scholar]
- 27.Powers, M. A., and D. J. Forbes. 1994. Cytosolic factors in nuclear transport: what's importin? Cell 79:931-934. [DOI] [PubMed] [Google Scholar]
- 28.Ren, X., J. S. Harms, and G. A. Splitter. 2001. Bovine herpesvirus 1 tegument protein VP22 interacts with histones, and the carboxyl terminus of VP22 is required for nuclear localization. J. Virol. 75:8251-8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandri-Goldin, R. M., and G. E. Mendoza. 1992. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 6:848-863. [DOI] [PubMed] [Google Scholar]
- 31.Turin, L., S. Russo, and G. Poli. 1999. BHV-1: new molecular approaches to control a common and widespread infection. Mol. Med. 5:261-284. [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng, C., L. A. Babiuk, and S. van Drunen Littel-van den Hurk. 2005. Bovine herpesvirus-1 VP22 enhances the efficacy of a DNA vaccine in cattle. J. Virol. 79:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng, C., R. Brownlie, L. A. Babiuk, and S. van Drunen Littel-van den Hurk. 2004. Characterization of nuclear localization and export signals of the major tegument protein VP8 of bovine herpesvirus-1. Virology 324:327-339. [DOI] [PubMed] [Google Scholar]