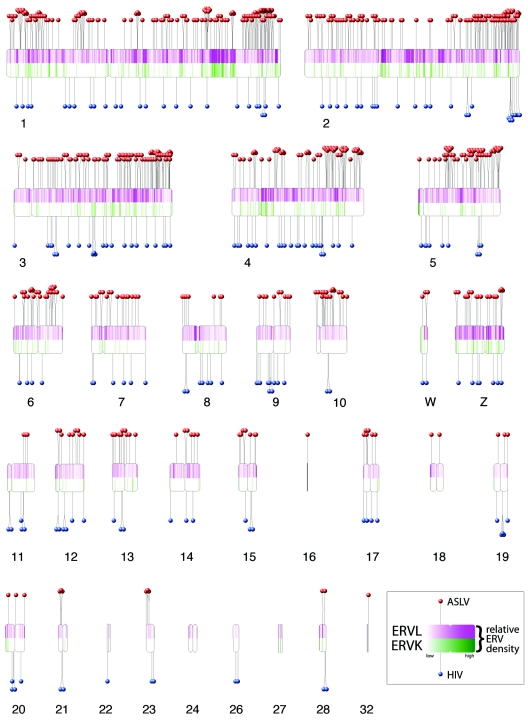
FIG. 1.
De novo integration targeting in the chicken genome and its relationship to ERVs. The chicken chromosomes are shown numbered (macrochromosomes 1 to 5, intermediate chromosomes 6 to 10, microchromosomes 11 to 32, and sex chromosomes W and Z). Note that, due to the incomplete status of the draft chicken genome sequence, some of the microchromosomes are not represented. Each de novo integration site is shown as a “lollipop” (ASLV, red; HIV, blue). Endogenous ERVL sequences are shown by the purple shading in the upper half of each chromosome, and ERVK is shown below by green. The most intense purple shading represents 85 ERVL integrations per 250 kbp; the most intense green shading represents 12 ERVK integrations per 250 kbp. Centromere locations are denoted by chromosomal indentations. The centromere positions for chromosomes 6, 9, 13 to 16, 18 to 22, 24, 27, and 32 are currently unavailable and have been arbitrarily placed at the chromosomal midpoints. The software used to draw the ideogram was obtained and adapted from http://www.uni-essen.de/∼bt0756/cc/.