FIG. 1.
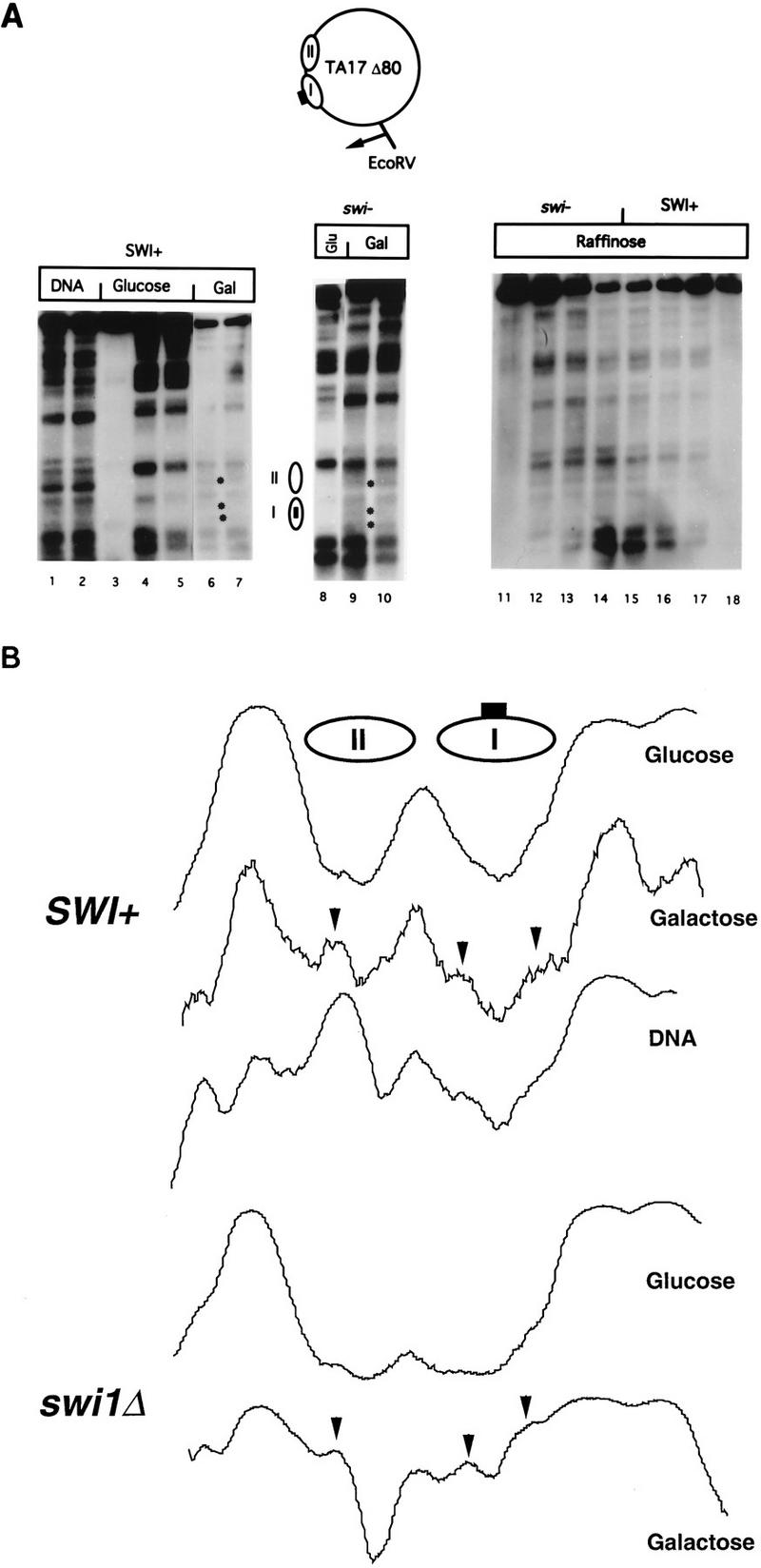
Perturbation of positioned nucleosomes in the yeast episome TA17Δ80 by GAL4 binding in SWI+ and swi1Δ yeast cells. (A) MNase cleavage sites in chromatin from SWI+ and swi1Δ cells grown in glucose, raffinose, or galactose medium were mapped relative to the EcoRV site as indicated. Samples were digested with MNase at 4 U/ml (lane 1), 10 U/ml (lane 2), 0 U/ml (lanes 3, 11, and 18), 2 U/ml (lanes 4, 6, 12, and 17), 5 U/ml (lanes 5, 7, 8, 10, 13, and 16), or 20 U/ml (lanes 9, 14, and 15). Bands cleaved in chromatin from cells grown in galactose are marked by asterisks to the right of lanes 6 and 9. Locations of nucleosomes I and II in cells grown in glucose are indicated; the box in nucleosome I represents the GAL4-binding site. (B) Densitometric scans of MNase cleavage patterns in the vicinity of nucleosomes I and II. Scans represent, in descending order, lanes 4, 6, 1, 8, and 10 in panel A. Arrowheads indicate cleavage sites induced in galactose. Note that the cleavage sites in the region of nucleosome I, which are cleaved weakly in chromatin from cells grown in galactose, are also weakly cut in naked DNA.
