FIG. 2.
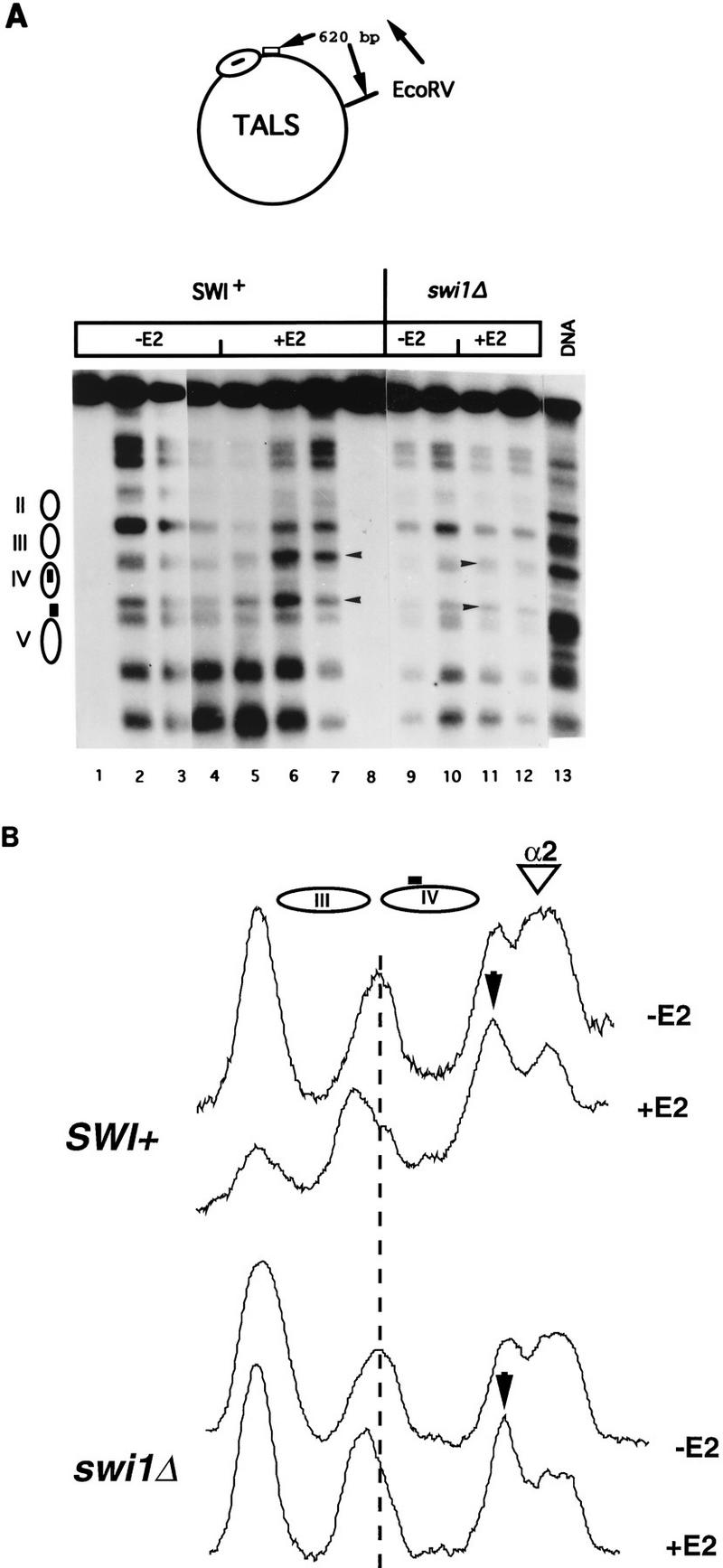
(A) Perturbation of TALS chromatin by the GAL4·ER·VP16 activation domain in SWI+ and swi1Δ yeast haploid α cells. MNase cleavage sites in cells harboring TALS and expressing GAL4· ER·VP16, either incubated for 4 h with 0.1 μM β-estradiol (+E2 lanes) or not (−E2 lanes), were mapped relative to the EcoRV site as indicated. Samples were digested with MNase at 0 U/ml (lanes 1 and 8), 5 U/ml (lanes 2 and 7), 20 U/ml (lanes 3, 6, 9, and 12), 50 U/ml (lanes 4, 5, 10, and 11), or 10 U/ml (lane 13). Control samples (0 U/ml) for the swi1Δ samples were identical in appearance to those shown for the SWI+ samples (data not shown). Arrowheads indicate bands cleaved preferentially under activating (+E2) conditions. The locations of nucleosomes II to V are indicated; the box in nucleosome IV represents the GAL4-binding site, and the box between nucleosomes IV and V represents the α2/MCM1 operator. (B) Densitometric scans of MNase cleavage patterns from lanes 4, 5, 10, and 11. The arrowhead indicates the lower of the two hormone-induced enhanced cleavage sites seen in the panel A, and the dotted line allows visualization of the slight shift in position of the cleavage site corresponding to the upper arrowhead in panel A.
