FIG. 4.
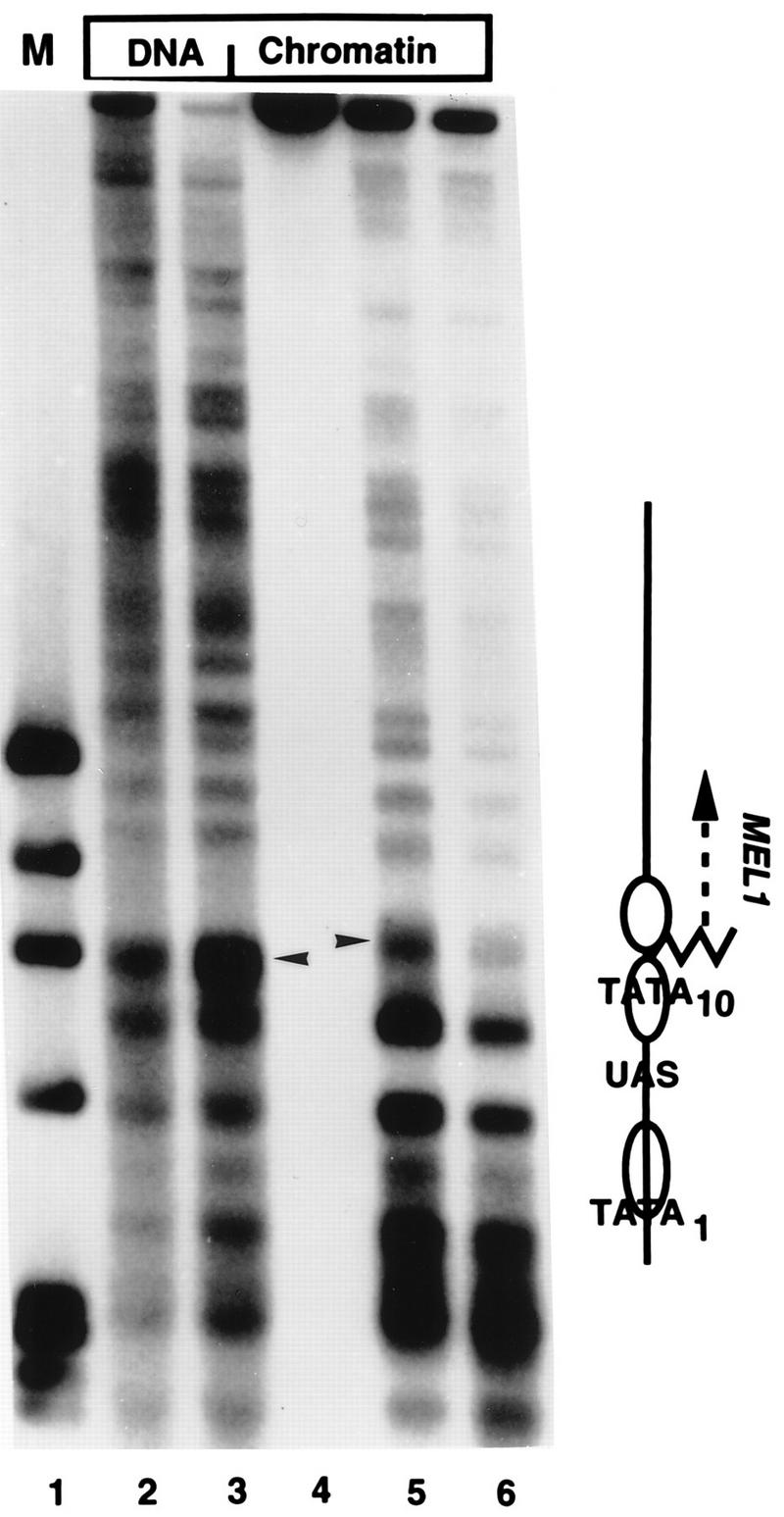
Indirect end-label analysis of the chromatin structure of the GAL10-MEL1 gene fusion. MNase cleavage sites in naked DNA or chromatin, as indicated, were mapped relative to a SalI site 830 bp upstream of the GAL10 TATA element, on the GAL1 side of the promoter in plasmid pBM150SKMEL1. Samples were digested with 4 (lane 2), 10 (lane 3), 0 (lane 4), 150 (lane 5) or 300 (lane 6) U of MNase per ml. Note the strong cleavage in naked DNA (arrowhead, lanes 2 and 3), which is protected in chromatin; a new cleavage (arrowhead, lanes 5 and 6) is present slightly higher on the gel, corresponding to the nucleosome-sized protected region containing the GAL10 TATA element. The overall pattern of cleavages in the vicinity of the UAS and GAL10 TATA is identical to that seen in the endogenous GAL1-10 promoter (data not shown). The locations of relevant promoter elements are indicated on the right; the jagged line represents the site of the fusion between the GAL10 promoter and the MEL1 coding sequence. Lane 1 contains ΦX/HaeIII markers.
