Abstract
The estrogen receptor (ER) is a ligand-dependent transcription factor that regulates the expression of estrogen-responsive genes. ER-mediated transcriptional changes are brought about by interaction of the ER with the estrogen response element (ERE). In this study, we examined the interaction of the Xenopus laevis ER DNA binding domain (DBD) and the intact ER with the X. laevis vitellogenin A2 ERE and the human pS2 ERE. Using gel mobility shift, DNase I footprinting, and methylation interference assays, we demonstrated that the DBD bound only as a dimer to the A2 ERE. However, the DBD bound as a monomer to the consensus pS2 ERE half site at lower DBD concentrations and then as a homodimer to the consensus and imperfect pS2 ERE half site at higher DBD concentrations. Antibody supershift experiments carried out with partially purified, yeast-expressed full-length ER demonstrated that three ER-specific antibodies interacted differentially with A2 and pS2 ERE-bound ER, indicating that receptor epitopes were differentially exposed. Furthermore, partial digestion of the A2 and pS2 ERE-bound ER with chymotrypsin or trypsin produced distinct protease cleavage patterns. Taken together, these data provide evidence that differential interaction of the DBD with the A2 and pS2 EREs brings about global changes in ER conformation. The conformational changes in ER induced by individual ERE sequences could lead to association of the receptor with different transcription factors and assist in the differential modulation of estrogen-responsive genes in target cells.
Estrogen is a hormone of central importance in regulating the development, growth, and maintenance of reproductive tissues. Estrogen’s actions are mediated by the intracellular estrogen receptor (ER), which interacts with estrogen response elements (EREs) present in target genes to bring about changes in transcription. Although the ER-ERE interaction plays a crucial role in regulating gene expression, the mechanisms by which this interaction leads to changes in transcription are unclear.
A number of thermodynamic and structural studies have demonstrated that specific contacts between protein and DNA are often accompanied by conformational changes in protein, DNA, or both (1, 9, 31, 39, 42, 48). These findings have led to the hypothesis that DNA can act as an allosteric modulator of protein conformation in a number of different systems (9, 39). For example, basic regions of leucine zipper proteins are poorly ordered in solution but are induced to form α-helical structures upon binding to DNA (31, 43). Nuclear factor NF-κB p50 subunits form chymotrypsin-resistant homodimers that serve as powerful transcriptional activators when bound to some recognition sequences (10). However, when bound to other recognition sequences, the same p50 subunits are degraded by chymotrypsin and are poor transcription activators. This differential sensitivity to protease digestion implies that homodimer conformations differ and that conformational variations can lead to differences in transcription activation.
The ER DNA binding domain (DBD) and the glucocorticoid receptor DBD undergo conformational changes on binding to their cognate hormone response elements. X-ray crystallographic studies demonstrate that local DBD regions, which are unfolded in solution, assume more ordered structures when bound to DNA (15, 21, 36, 37). In addition, crystallographic analysis of the ER DBD bound to the vitellogenin B1 ERE2 (AGTCAnnnTGACC [50]), which differs from the vitellogenin A2 ERE (GGTCAnnnTGACC [16]) by a single base pair (underlined), has demonstrated that the substitution of an adenine for a guanine in the 5′ half site causes the rearrangement of a lysine side chain, disruption of a salt bridge between lysine and glutamic acid residues, and destruction of a hydrogen bond with the guanine residue (38). When the DBD is bound to the vitellogenin B1 ERE2, the lysine residue accommodates the nucleotide substitution by forming hydrogen bonds with a nearby tyrosine residue and the substituted adenine residue. Thus, the change of one nucleotide requires the formation of a new and different interconnected hydrogen bond network and implies that each ERE sequence may induce unique conformational changes in DBD structure.
At this point, it is uncertain whether changes in DBD conformation can be transmitted to other receptor regions and thereby alter receptor function. Starr et al. (41) have provided evidence that mutation of a single amino acid in the glucocorticoid receptor DBD induces conformational changes in a transcription activation domain of the receptor. However, other studies have demonstrated that ER DNA and ligand binding domains function as independent entities, which can be fused to heterologous units and still effectively activate transcription (12, 20, 45, 51).
A number of laboratories have demonstrated that EREs with imperfect ERE half sites are weaker transcriptional activators than the A2 ERE (6, 22, 32). Interestingly, we recently demonstrated that the orientation of a consensus or an imperfect ERE relative to the TATA sequence can have profound effects on the expression of an estrogen-responsive reporter plasmid (28). The A2 ERE maximally activates transcription when it is separated from the TATA sequence by 2.6 or 3.6 helical turns, whereas the pS2 ERE maximally activates transcription when it is separated from the TATA sequence by 3 helical turns. From these studies, we hypothesized that the ERE may act as an allosteric modulator of ER conformation and that these DNA-induced changes in ER conformation could in turn influence ER-protein interactions and lead to changes in transcription activation.
To determine if an ERE sequence could induce specific changes in receptor conformation, we have characterized the interaction of the ER DBD and the intact ER with the vitellogenin A2 and the pS2 ERE sequences. The Xenopus laevis vitellogenin A2 ERE is a perfectly palindromic, consensus ERE sequence (GGTCAnnnTGACC [16]) and differs from the human pS2 ERE in the 3′ half site by one base pair (GGTCAnnnTGGCC [30]). We detect differences in the interaction of the purified ER DBD with the A2 and pS2 EREs in gel mobility shift, DNase I footprinting, and methylation interference assays. The differential interaction of ER-specific antibodies with A2 and pS2 ERE-bound ER implies that there are differences in ER conformation. Protease sensitivity assays provide further evidence that the conformations of the A2 and pS2 ERE-bound ER are distinct. We believe that these DNA-induced conformational changes in ER can form the basis for differential transcription of estrogen-responsive genes.
MATERIALS AND METHODS
Preparation of 32P-labeled DNA fragments, ER DBD, and ER.
For gel mobility shift assays, DNase I footprinting, and methylation interference experiments, 5 μg of circular permutation plasmids B3consERE and B3pS2ERE (28) were digested with EcoRV and HindIII to produce 278-bp ERE-containing DNA fragments containing the A2 and pS2 EREs, respectively, flanked by identical nucleotide sequence. To label the coding strand, the ERE-containing DNA fragments were combined with 50 mM Tris (pH 8.0), 10 mM MgCl2, 50 mM NaCl, 25 pmol (150 μCi) of [α-32P]dATP, 25 pmol (150 μCi) of [α-32P]dGTP, 140 μM dTTP, 140 μM dCTP, and 1 U of Klenow DNA polymerase in a final volume of 40 μl. After 20 min at room temperature, 140 μM dATP and 140 μM dGTP were added to the samples, and the reaction mixture was incubated for another 5 min at room temperature. DNA fragments were fractionated on a 5% acrylamide gel, excised, isolated by electroelution, precipitated, and resuspended in TE (10 mM Tris [pH 8.0], 1 mM EDTA). ERE-containing DNA fragments were also labeled on the noncoding strand and used in DNase I footprinting and methylation interference experiments. To label the noncoding strand, plasmids B3consERE and B3pS2ERE were cut with EcoRV and NheI to produce 388-bp ERE-containing DNA fragments. The fragments were filled in on the noncoding strand as described above except that 25 pmol (150 μCi) of [α-32P]dCTP, 25 pmol (150 μCi) of [α-32P]dTTP, 140 μM dATP, and 140 μM dGTP were used. After 20 min, 140 μM dCTP and 140 μM dTTP were added to the reaction mixture. 32P-labeled probes were fractionated on an acrylamide gel and electroeluted as described for the coding strand.
For the antibody supershift experiments, 5 μg of each of plasmids B3consERE and B3pS2ERE was cut with HindIII and 32P labeled as described above for the coding strand. The 425-bp, end-labeled, ERE-containing DNA fragments were gel purified on a 5% acrylamide gel, excised, electroeluted, precipitated, and resuspended in TE.
For protease sensitivity experiments, 5-μg aliquots of plasmids B3consERE and B3pS2ERE were cut with EcoRI and BamHI to produce 55-bp ERE-containing DNA fragments. The fragments were gel purified and labeled as described above except that 49.5 pmol (300 μCi) of [α-32P]dATP and 16.5 pmol (100 μCi) of [α-32P]dGTP were used. The probes were gel purified a second time on a 5% acrylamide gel, excised, electroeluted, precipitated, and resuspended in TE.
The expression and purification of the 111-amino-acid X. laevis ER DBD (amino acids 171 to 281) and the partially purified yeast-expressed human ER have been described elsewhere (27, 28). These studies were carried out exclusively with the ERα DBD and full-length receptor, not the recently discovered ERβ (18).
Gel mobility shift assays.
Gel mobility shift assays were carried out as previously described (29). Briefly, EcoRV/HindIII 32P-labeled DNA fragments (0.05 to 0.1 pmol) containing the A2 ERE were combined with 0 to 0.37 pmol of purified DBD in binding reaction buffer (15 mM Tris [pH 7.9], 0.2 mM EDTA, 10% glycerol, 4 mM dithiothreitol) with 80 mM KCl and 50 ng of poly(dI-dC) to a final volume of 20 μl. The DBD-DNA mixture was incubated for 15 min at room temperature and then fractionated on an 8% low-ionic-strength acrylamide gel. 32P-labeled DNA fragments containing the pS2 ERE were identically processed except that 0 to 1.83 pmol of purified DBD were used in the binding reactions.
DNase I footprinting.
EcoRV/HindIII-digested A2 ERE-containing DNA fragments (0.5 to 1.0 pmol), which had been labeled on the coding strand, were combined with 0 to 7.34 pmol of purified DBD in binding reaction buffer with 80 mM KCl, 50 ng of poly(dI-dC), 1.25 mM MgCl2, and 0.5 mM CaCl2 to a final volume of 20 μl. DNA fragments containing the pS2 ERE were identically processed except that 0 to 36.7 pmol of purified DBD was used. Ovalbumin was also included in each reaction so that the total protein concentration was 2.5 μg. The binding reaction mixtures were incubated for 15 min at room temperature. Then 0.4 U RQ1 RNase-free DNase I (Promega, Madison, Wis.) was added in the absence of the DBD, and 0.8 U of DNase I was added to reactions containing the DBD. The samples were cleaved for 1.5 or 2.5 min, respectively, after which digestion was terminated by addition of 20 μl of DNase I stop solution (200 mM NaCl, 1% sodium dodecyl sulfate, 30 mM EDTA). The DNA was extracted with phenol-chloroform, precipitated, washed twice with 70% ethanol, and dried. The A2 and pS2 ERE-containing DNA fragments were resuspended in loading buffer, incubated at 90°C for 1.5 min, and electrophoresed on an 8% sequencing gel. The gel was dried and visualized by autoradiography.
The protection of each A2 and pS2 ERE half site was quantitated by using a PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.). Each lane was normalized to account for unequal loading, and then the level of radioactivity in each ERE half site was quantitated before and after addition of increasing amounts of DBD. The level of protection, which is expressed as the percentage of cleaved DNA, was calculated by determining the amount of cleaved DNA in each ERE half site in the presence of DBD relative to the amount of cleaved DNA in each ERE half site in the absence of DBD.
Methylation interference.
EcoRV/HindIII-digested, end-labeled DNA fragments (8 to 10 pmol; 106 cpm) were methylated in 211 μl of DMS (dimethyl sulfate) buffer (50 mM sodium cacodylate [pH 8.0], 1 mM EDTA) with 0.5% DMS. After 3 min, the reaction was terminated with 50 μl of DMS stop solution (1.5 M sodium acetate [pH 7.0], 1 M β-mercaptoethanol, 100 μg of tRNA per ml) and 750 μl of cold ethanol. The modified DNA was precipitated twice and resuspended in TE. Then 1.5 to 3.0 pmol of methylated A2 or pS2 ERE-containing probe was combined with 3.7 or 7.4 pmol of purified DBD, respectively, in binding reaction buffer with 80 mM KCl and 50 ng of poly(dI-dC). The 20-μl reaction mixture was fractionated on an 8% nondenaturing polyacrylamide gel. The free probe and protein-DNA complexes were detected by autoradiography of the wet gel and excised. The modified DNA was isolated by electroelution, precipitated, and then cleaved for 30 min with 10% piperidine at 90°C. The piperidine solution was evaporated, and the modified DNA was resuspended in 30 μl of water, lyophilized, resuspended in 20 μl of water, and lyophilized. The A2 and pS2 ERE-containing DNA fragments were resuspended in loading buffer, incubated at 90°C for 1.5 min, and electrophoresed on an 8% sequencing gel. The gel was dried and visualized by autoradiography.
Antibody supershifts.
Monoclonal antibody P1A3 was made against purified X. laevis ER DBD at the Immunological Resource Center, University of Illinois at Urbana-Champaign. The production of antibodies ER 21, H226, D547, H222, and D75 has been described previously (4, 13). Polyclonal antibodies ER6 and ER1 and monoclonal antibody h151 were provided by Robin Fuchs-Young (M. D. Anderson Cancer Center, University of Texas, Smithville) and Dean Edwards (University of Colorado, Denver), respectively.
Gel mobility supershift assays were carried out with partially purified, yeast-expressed human ER (28). For these assays, 0.05 to 0.1 pmol of the EcoRV/HindIII-digested, end-labeled DNA fragments containing the A2 or pS2 ERE were combined with 285 or 570 fmol of ER in binding reaction buffer with 10 μg of bovine serum albumin, 1 μg of poly(dI-dC), 20 mM KCl, 50 μM ZnCl2, and 10−7 M 17β-estradiol (E2). The reaction mixtures were incubated for 10 min at room temperature before one of the indicated ER-specific antibodies was added to the A2 or pS2 ERE-containing samples. After 5 min at room temperature, the protein-DNA complexes were fractionated for 4 h at 300 V on a nondenaturing 8% acrylamide gel and processed as described above. The amount of free and bound DNA was determined with a PhosphorImager and ImageQuant software.
Partial ER proteolysis.
The 55-bp, 32P-labeled DNA fragments (0.05 to 0.1 pmol) containing either the A2 or pS2 ERE were combined with 285 or 570 fmol of ER, respectively, as described above. After a 10-min incubation, 0, 0.05, 0.5, 1.25, 2.5, 3.75, or 5 ng of chymotrypsin (Sigma, St. Louis, Mo.) was added to the A2 and pS2 ERE-containing reaction mixtures. The samples were incubated for an additional 10 min and loaded onto a running, 8% nondenaturing acrylamide gel. The gel was electrophoresed for 2 h at 300 V, dried, and visualized by autoradiography. A2 or pS2 ERE-ER complexes were also exposed to trypsin cleavage and processed similarly except that 0, 0.05, 0.5, 1.25, 2.5, 3.75, or 5 ng of trypsin (Worthington Biochemical Corporation, Freehold, N.J.) was added to each sample.
RESULTS
To be certain that the ER-ERE interaction was not influenced by other proteins, we began our investigations by using highly purified preparations of X. laevis ER DBD. There are several advantages to using the DBD. First, it is easily expressed in bacteria and can be highly purified in a two-step chromatographic procedure (27). Second, the DBD retains many of the characteristics of the intact receptor including specific interaction with the ERE, differential binding to EREs that deviate from the consensus sequence, and activation of an estrogen-responsive reporter construct (6, 27). Third, the DBD structure has been defined in detail by nuclear magnetic resonance and X-ray crystallographic techniques (36–38). Fourth, because the amino acid sequence of steroid hormone receptor DBDs is highly conserved, delineating how one DBD interacts with its cognate response element may also help to delineate how other members of the nuclear receptor superfamily activate transcription.
Differential interaction of the DBD with A2 and pS2 EREs is detected in gel mobility shift assays.
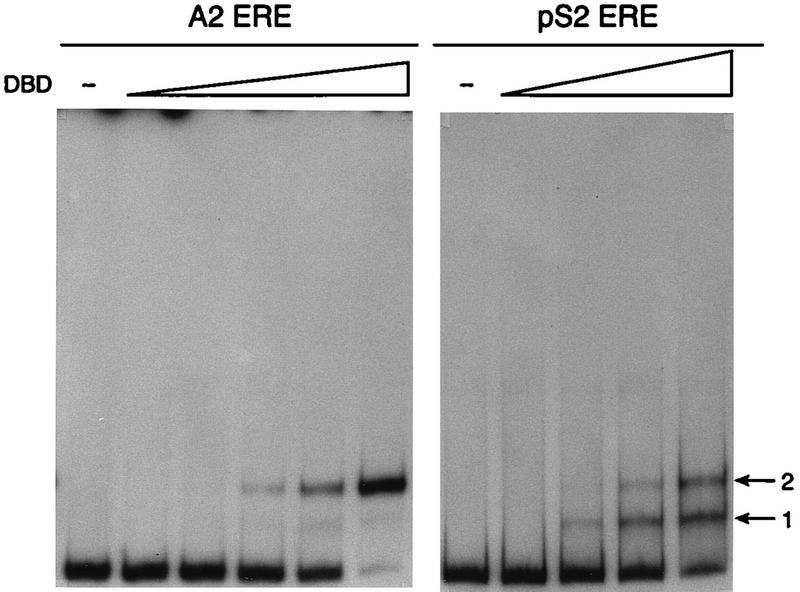
To begin characterizing the DBD-ERE interaction, gel mobility shift assays were carried out. 32P-labeled DNA fragments containing the A2 ERE or the pS2 ERE were combined with increasing amounts of purified DBD and fractionated on nondenaturing polyacrylamide gels. The DBD formed a single complex with the A2 ERE regardless of DBD concentration, suggesting that the DBD occupied both of the consensus ERE half sites (Fig. 1). These results are consistent with previous X-ray crystallographic and gel mobility shift assays which demonstrate that even at extremely low DBD concentrations, the DBD bound as a dimer to a consensus ERE sequence (27, 36, 38). In contrast to our findings with the A2 ERE, DNA fragments containing the pS2 ERE formed two complexes with the DBD. Complex 2 had the same mobility as the single complex formed with the A2 ERE, indicating that the DBD was probably binding as a dimer to the pS2 ERE (Fig. 1, arrow 2). Complex 1 migrated more rapidly than complex 2 and probably represents one DBD monomer interacting with the pS2 ERE-containing DNA fragments (Fig. 1, arrow 1). The disappearance of complex 1 and the appearance of complex 2 with increasing DBD concentration supports the idea that a monomer-to-dimer transition was occurring with the pS2 ERE. Although the DBD bound to both the A2 and pS2 EREs, significantly lower levels of DBD were required for occupation of the consensus A2 ERE than for occupation of the imperfect pS2 ERE. This was not surprising since we have previously demonstrated that the affinity of the intact receptor is twofold lower for the pS2 ERE than for the A2 ERE (28).
FIG. 1.
The DBD forms one complex with the A2 ERE but forms two complexes with the pS2 ERE. Increasing concentrations of purified DBD (0, 0.007, 0.03, 0.07, 0.18, or 0.36 pmol for the A2 ERE; 0, 0.07, 0.36, 0.73, or 1.83 pmol for the pS2 ERE) were incubated with 32P-labeled A2 or pS2 ERE-containing DNA fragments as described in Materials and Methods. The binding reactions were fractionated on a nondenaturing acrylamide gel, and the gel was dried and subjected to autoradiography. Complexes 1 and 2, formed between the DBD and A2 or pS2 EREs, are indicated to the right.
DNase I footprinting demonstrates that the DBD dimer interacts with the A2 ERE but that both the DBD monomer and dimer interact with the pS2 ERE.
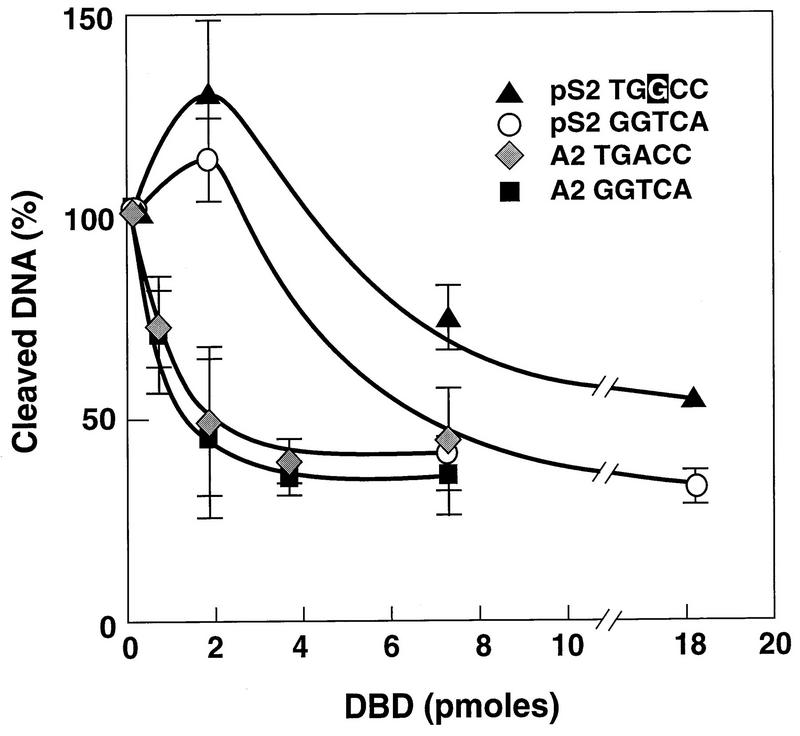
To determine if the complexes formed in the gel shift assays corresponded to DBD monomer and dimer binding and to further characterize the interaction of the DBD with the A2 and pS2 EREs, DNase I footprinting was carried out. This technique utilizes the nonspecific cleavage properties of DNase I to identify DNA regions that are protected by proteins. DNA fragments containing the A2 ERE or the pS2 ERE were 32P-labeled on the coding strand and then combined with increasing amounts of purified DBD. The reactions were subjected to DNase I digestion, and the resulting cleavage products were separated on a sequencing gel. As seen in Fig. 2, the DBD interacted only with the region of the DNA fragments that included either the A2 ERE or the pS2 ERE. Although the areas of protection were similar for the A2 and pS2 EREs, there were distinguishable differences in the pattern of cleavage. Quantitative analysis of the A2 and pS2 ERE half sites demonstrated that both A2 ERE half sites were equally protected regardless of protein concentration (Fig. 3). These findings indicated that the DBD bound to each ERE half site with equal affinity and confirmed that only the DBD dimer bound to the A2 ERE. The pS2 ERE half sites, however, were differentially protected with the consensus pS2 ERE half site (GGTCA), requiring lower DBD concentrations for protection than the imperfect pS2 ERE half site (TGGCC). Thus, the DBD bound to the pS2 ERE as a monomer at lower DBD concentrations and as a dimer at higher DBD concentrations. It should be noted, however, that higher DBD concentrations were required for protection of the pS2 ERE than for protection of the A2 ERE (Fig. 2 and 3). Interestingly, hypersensitive sites were observed at the 3′ ends of both the A2 and pS2 ERE footprints (Fig. 2, ∗).
FIG. 2.
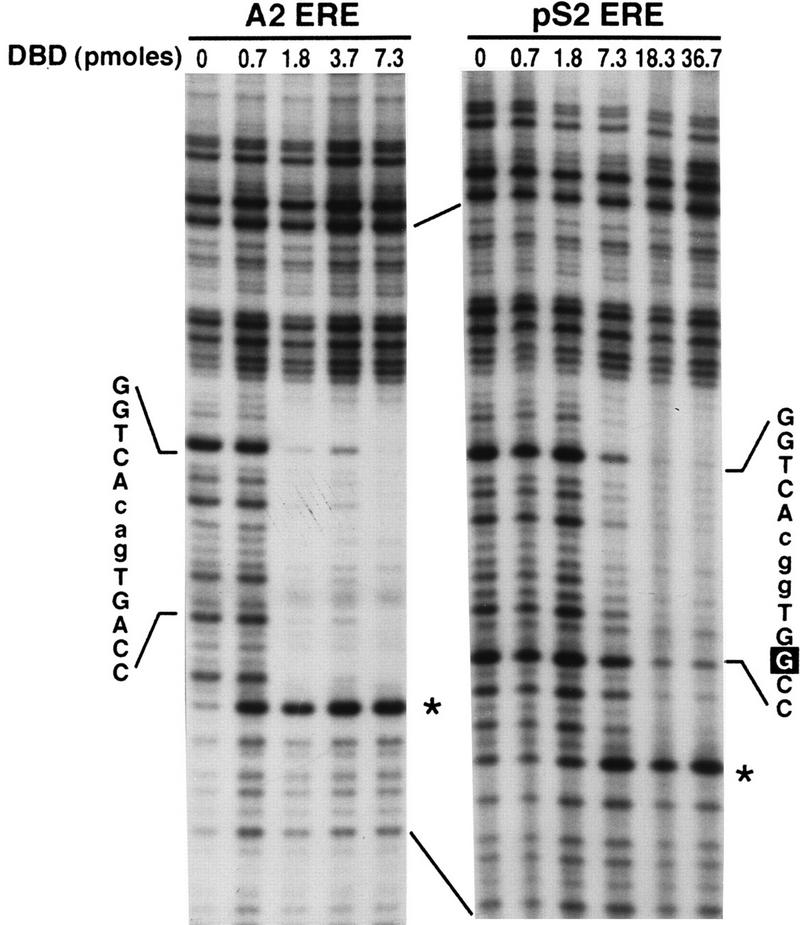
DNase I footprinting defines regions of the coding strand that are involved in DBD binding. Increasing concentrations of purified DBD were incubated with A2 or pS2 ERE-containing DNA fragments which had been labeled on the coding strand. The binding reactions were subjected to limited DNase I digestion, and the cleaved DNA was fractionated on a denaturing acrylamide gel. The gel was dried and subjected to autoradiography. The positions and sequences of the A2 ERE and the pS2 ERE and DNase I-hypersensitive sites (∗) are indicated.
FIG. 3.
Lower DBD concentrations are required to protect the A2 ERE than the pS2 ERE. The level of A2 and pS2 ERE half site protection on the coding strand is expressed as the percentage of cleaved DNA and was calculated by determining the amount of cleaved DNA in each ERE half site in the absence of DBD relative to the amount of cleaved DNA in each ERE half site in the presence of DBD. Each point represents the mean ± standard error of the mean from three to four independent experiments.
DNA fragments containing the A2 or the pS2 ERE were also 32P-labeled on the noncoding strand and subjected to DNase I cleavage. Like the coding strand, the region protected on the noncoding strand included only the A2 or the pS2 ERE (Fig. 4), the A2 ERE half sites were equally protected, and lower DBD concentrations were required for protection of the pS2 ERE consensus half site than for protection of the imperfect pS2 ERE half site. Lower DBD concentrations were again required to protect the A2 ERE than the pS2 ERE. These data from the noncoding strand demonstrate that the DBD bound only as a dimer to the A2 ERE, while both the DBD monomer and dimer bound to the pS2 ERE.
FIG. 4.
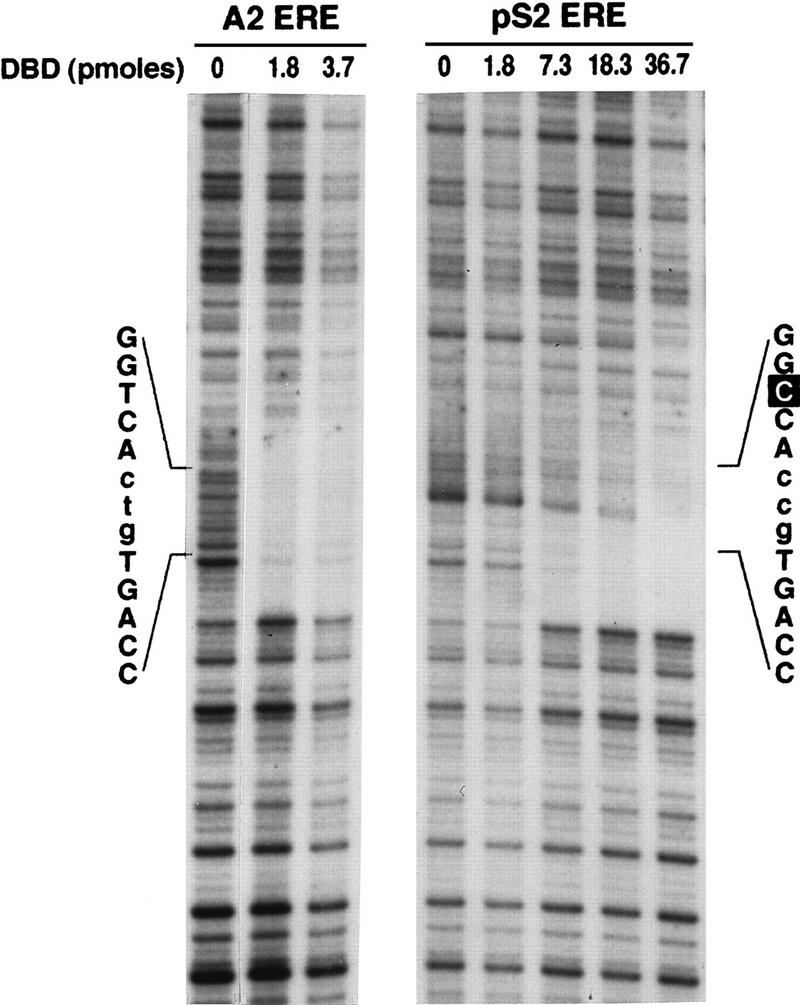
DNase I footprinting defines regions of the noncoding strand that are involved in DBD binding. Increasing concentrations of purified DBD were incubated with A2 or pS2 ERE-containing DNA fragments which had been labeled on the noncoding strand. The binding reaction was subjected to limited DNase I digestion, and the cleaved DNA was fractionated on a denaturing acrylamide gel. The gel was dried and subjected to autoradiography. The positions and sequences of the A2 and pS2 EREs are indicated.
Guanine residues in the A2 and pS2 EREs are required for DBD dimer binding, but only guanine residues in the pS2 ERE consensus half site are necessary for DBD monomer binding.
Because DNase I is a large globular protein, steric hindrance of this molecule with other proteins may result in an overestimation of the DNA region protected by bound proteins. Therefore, to more specifically define and compare the contacts between the DBD and the A2 and pS2 ERE sequences, methylation interference assays were carried out. This method of footprinting uses DMS, a small molecule, to modify guanine residues. The modified DNA is then incubated with a DNA binding protein that specifically interacts with a recognition sequence present in the DNA strand. Because methylation of guanine residues in a recognition sequence inhibits protein binding, guanine residues that are required for efficient protein-DNA interaction can be identified.
A2 ERE- or pS2 ERE-containing DNA fragments were modified by DMS treatment and combined with 5.5 or 29.3 pmol of purified DBD, respectively, so that approximately 50% of the DNA fragments were bound to the DBD. Free DNA and DBD-DNA complexes were fractionated on a nondenaturing acrylamide gel, isolated, cleaved, and resolved on a denaturing gel. Methylation of guanine residues in the A2 and pS2 ERE half sites strongly inhibited DBD binding, as indicated by the diminished intensity of the bands corresponding to these nucleotides (Fig. 5). Specific DBD binding required the participation of guanine residues (bold faced) in both half sites of the A2 ERE (GGTCAcagTGACC). Interaction of the DBD dimer with the pS2 ERE also required unmodified guanine residues in both the consensus and imperfect half sites (GGTCAcggTGGCC). These findings are consistent with X-ray crystallographic studies carried out with the ER DBD and methylation interference assays carried out with the full-length ER (17, 36). Of particular interest was the interaction of the DBD monomer with the pS2 ERE, which required only the participation of guanine residues in the consensus half site (Fig. 5). These data are in good agreement with our DNase I footprinting results (Fig. 2 to 4) and provide additional evidence that first the DBD binds as a monomer to the consensus pS2 ERE half site and then a second monomer binds to the imperfect pS2 ERE half site as DBD concentrations are increased.
FIG. 5.
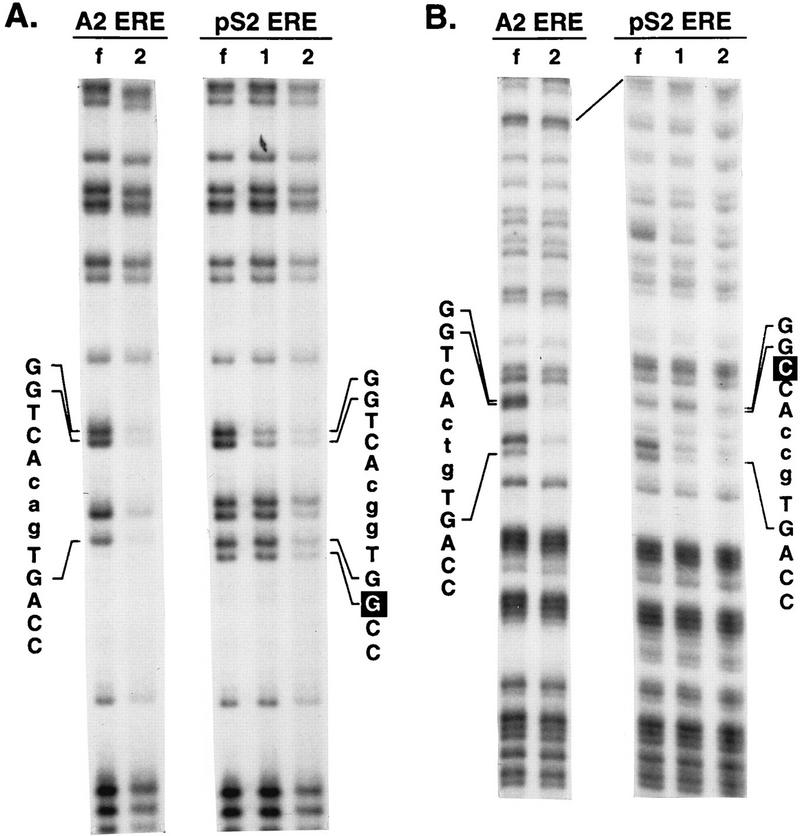
Methylation interference experiments delineate guanine residues required for efficient binding of the DBD to the A2 and pS2 EREs. A2 or pS2 ERE-containing DNA fragments which had been labeled on the coding (A) or noncoding (B) strand were modified with DMS. The modified DNA fragments were incubated with purified DBD and fractionated on a nondenaturing acrylamide gel. The free probe (lanes f), DBD-ERE complex 1 (lanes 1), and DBD-ERE complex 2 (lanes 2) were detected by autoradiography. The DNA from each band was isolated, cleaved, fractionated on a denaturing gel, and visualized by autoradiography. The positions and sequences of the A2 and pS2 EREs are indicated.
Differences in ER epitope availability are detected in antibody supershift experiments when the receptor is bound to the A2 or the pS2 ERE.
The gel shift and footprinting experiments established that the DBD interacted differently with the A2 and pS2 EREs but did not provide direct evidence that DBD conformation was different when bound to these two EREs. We reasoned that subtle changes in DBD structure might be translated to other ER regions, resulting in more global conformational changes in the intact receptor. Therefore, monoclonal and polyclonal antibodies directed against several ER regions (Fig. 6A) were used in antibody supershift experiments to determine if differences in epitope availability could be detected when the ER was bound to the A2 or pS2 ERE. Partially purified, yeast-expressed ER was combined with 32P-labeled A2 or pS2 ERE-containing DNA fragments. Antibodies directed against different ER epitopes were then added to the binding reaction mixtures, and the resulting complexes were fractionated on nondenaturing polyacrylamide gels. The level of each receptor-DNA complex was quantitated so that the effect of each antibody on the A2 and the pS2 ERE-ER complex formation could be assessed. The most striking difference in epitope availability was observed with monoclonal antibody P1A3, which was made against purified X. laevis DBD. P1A3 enhanced the ER-A2 ERE complex formation approximately sixfold (Fig. 6B; compare lanes 1 and 7) and strongly inhibited ER-pS2 ERE complex formation (compare lanes 2 and 8) but failed to supershift either the A2 or pS2 ERE-ER complex. Two other antibodies also discriminated between the pS2 and A2 ERE-bound ER. ER21 and D75, which are directed against the amino and carboxy termini of the receptor, respectively, did not alter the supershifted ER-A2 ERE complex formation but decreased formation of the ER-pS2 ERE complex (Fig. 6B; compare lane 2 with lanes 4 and 20). The decreased pS2 ERE-ER complex formation was more apparent when increased amounts of receptor were included in the binding reaction (Fig. 6C). The other antibodies tested (H226, ER6, ER1, D547, h151, and H222) supershifted both the A2 and pS2 ERE-containing complexes in a similar manner. The differential interaction of three ER-specific antibodies with A2 and pS2 ERE-bound ER implied that there were differences in ER epitope availability not only in the DBD but also in the amino and carboxy termini of the receptor.
FIG. 6.
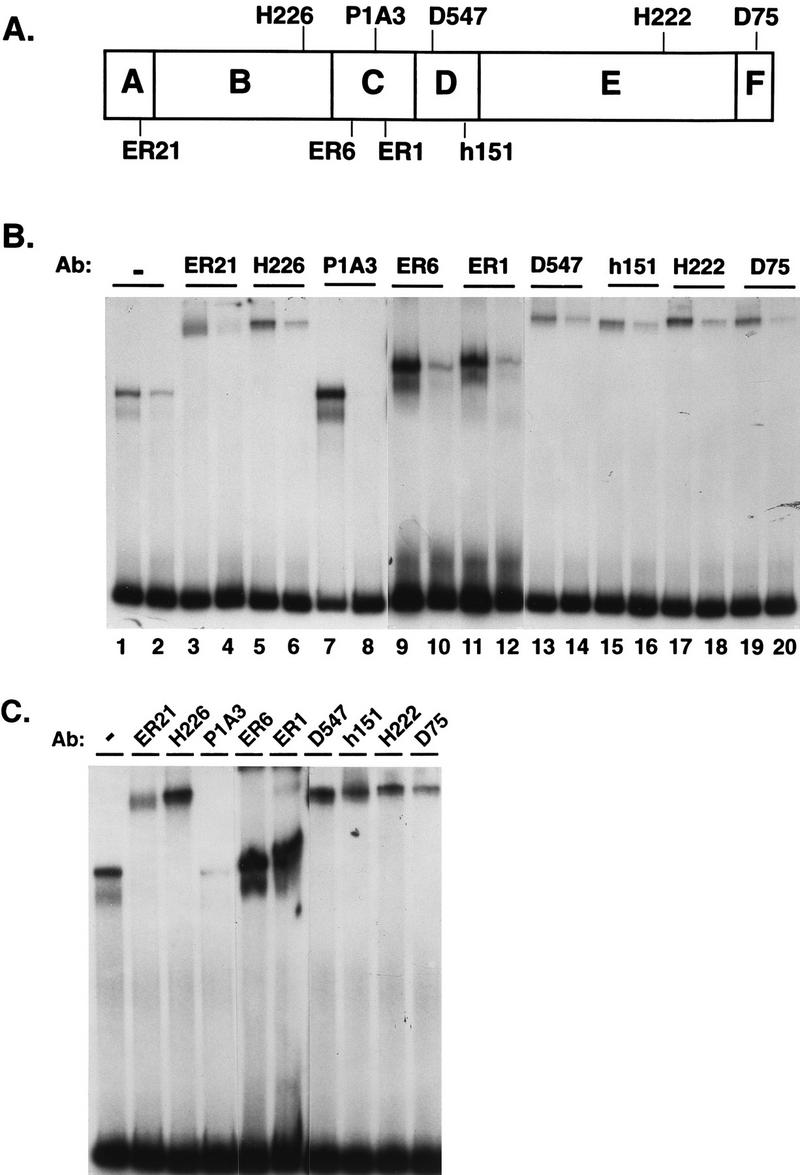
Antibodies to various ER epitopes can detect differences in conformation of the A2 and pS2 ERE-bound receptor. (A) Schematic representation of the epitopes for ER-specific antibodies used. (B) Partially purified, yeast-expressed ER (285 fmol) was incubated with A2 ERE-containing DNA fragments (odd-numbered lanes) or pS2 ERE-containing DNA fragments (even-numbered lanes). After a short incubation, antibodies (Ab) were added to the binding reactions as indicated and the complexes were fractionated on a nondenaturing acrylamide gel. The complexed DNA and free probe were visualized by autoradiography. (C) Partially purified, yeast-expressed ER (570 fmol) was incubated with pS2 ERE-containing DNA fragments. Samples were processed as for panel B.
Sequence-mediated changes in ER conformation are detected after limited protease digestion of the A2 and pS2 ERE-bound ER.
The antibody supershift experiments provided evidence that ER epitopes were differentially exposed when the receptor was bound to the A2 and pS2 EREs and therefore that differences in receptor conformation may exist. To more directly assess possible differences in receptor conformation, protease sensitivity assays were carried out with A2 and pS2 ERE-bound ER. This assay utilizes limited proteolysis of a DNA-bound protein to produce a pattern of digestion based on amino acid accessibility and provides information about native protein conformation (35, 44). 32P-labeled DNA fragments containing the A2 or pS2 EREs were combined with 285 or 570 fmol of partially purified yeast-expressed ER, respectively. This twofold difference in ER concentration was used to account for the lower binding affinity of the intact ER for the pS2 ERE (28). The protein was then subjected to limited proteolysis by exposure to increasing concentrations of chymotrypsin, and the resulting complexes were fractionated on nondenaturing polyacrylamide gels. The differences in the digestion patterns observed with the A2 ERE-bound ER and the pS2 ERE-bound ER were striking (Fig. 7A). Limited digestion of the A2 ERE-bound ER produced a larger stable ER-DNA complex (C3) than was observed with the pS2 ERE-bound ER after chymotrypsin treatment (C5). The numbers of intermediate ER-DNA complexes observed with A2 and pS2 ERE-bound ER were also quite distinct. While chymotrypsin digestion of the A2 ERE-bound receptor produced several ER-DNA complexes of intermediate size (C1 to C5), digestion of the pS2 ERE-bound ER produced few intermediate-size ER-DNA complexes.
FIG. 7.
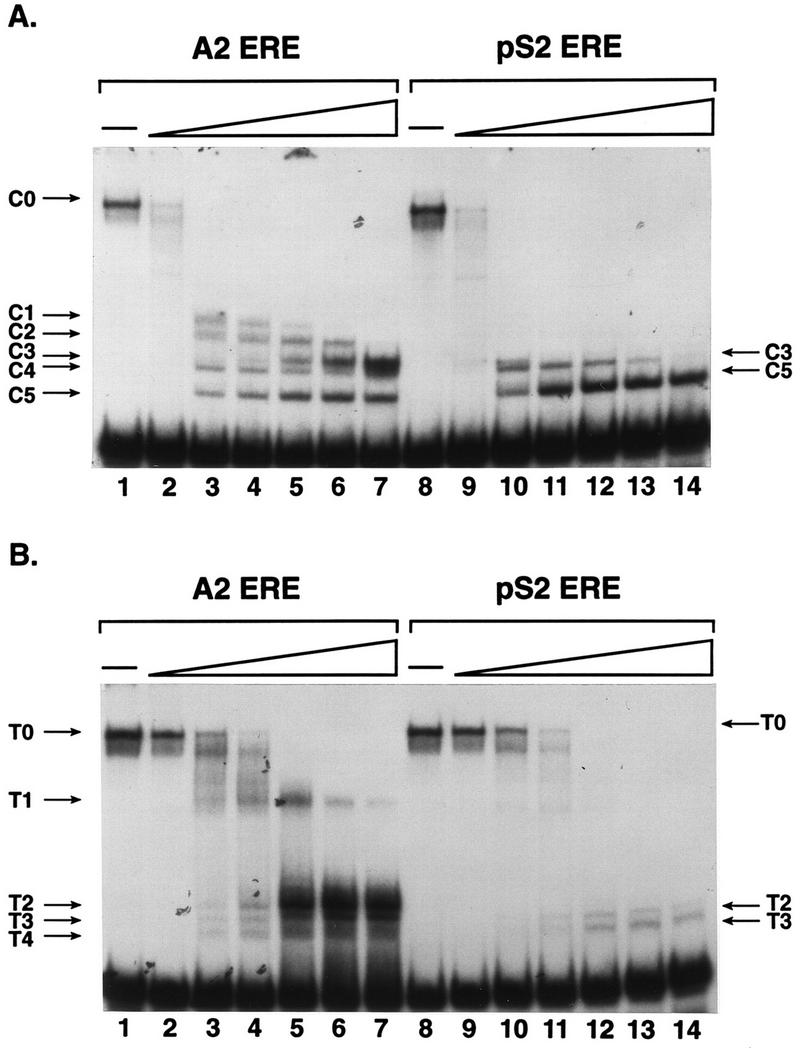
Distinct protease digestion patterns of A2 and pS2 ERE-bound ER provide evidence for ERE-mediated differences in receptor conformation. (A) Partially purified, estrogen-occupied ER was combined with A2 or pS2 ERE-containing DNA fragments. After a short incubation, 0, 0.05, 0.5, 1.25, 2.5, 3.75, or 5 ng of chymotrypsin was added to the binding reaction. ER-DNA complexes and free DNA were fractionated on a nondenaturing acrylamide gel, and the gel was dried and subjected to autoradiography. The undigested ER-DNA complex (C0) and ER-DNA complexes formed with chymotrypsin-proteolyzed receptor (C1 to C5) are indicated. (B) Partially purified ER and A2 or pS2 ERE-containing DNA fragments were combined as for panel A except that 0, 0.05, 0.5, 1.25, 2.5, 3.75, or 5 ng of trypsin was added to the binding reactions. The undigested ER-DNA complex (T0) and ER-DNA complexes formed with trypsin-proteolyzed receptor (T1 to T4) are indicated.
The difference in digestion patterns observed with these two EREs was not due to differences in ER or DNA concentrations, since different amounts of ER and DNA produced the same digestion pattern, nor was it due to a difference in chymotrypsin concentrations, since the same digestion patterns were produced at higher and lower chymotrypsin concentrations. Since we have observed similar digestion patterns with partially purified yeast-expressed ER and ER-containing nuclear extracts from estrogen-treated CHO-ER cells, the difference in digestion patterns did not result from the association of different proteins with the ER (data not shown). Furthermore, the difference in digestion patterns was not due to dissociation of the ER from the pS2 ERE and enhanced proteolysis of the free receptor, since the amount of ER-DNA complex observed at the highest chymotrypsin concentration was similar to the amount of ER-DNA complex observed in the absence of protease (compare lanes 8 and 14). Finally, the higher mobility complexes (C1 to C5) produced by chymotrypsin cleavage were due to specific cleavage of the protein and not degradation of DNA since the free DNA was not degraded as chymotrypsin levels increased. Digestion of the A2 and pS2 ERE-bound ER was done in parallel, and the results were completely reproducible.
Trypsin digestion also resulted in distinctly different cleavage patterns of the A2 and pS2 ERE-bound ER (Fig. 7B). Digestion of the A2 ERE-bound ER with trypsin produced several products (T1 to T4), with complex T2 being most stable at high trypsin concentrations (Fig. 7B, lanes 1 to 7). In contrast, digestion of the pS2 ERE-bound ER produced fewer trypsin products, with complex T3 being the most stable (lanes 8 to 14). The pS2 ERE-bound ER appeared to be particularly susceptible to trypsin cleavage as evidenced by the loss of ER-DNA complex at higher trypsin concentrations. Therefore, we believe that the different digestion patterns that we observed with the A2 and pS2 ERE-bound ER resulted from differences in receptor conformation and that the conformation was dictated by the ERE sequence.
DISCUSSION
This study focused on the differential interaction of the ER DBD and the intact receptor with A2 and pS2 EREs. The A2 ERE (GGTCAnnnTGACC [16]) differs from the pS2 ERE (GGTCAnnnTGGCC [30]) by a single base pair (underlined) in the 3′ half site. Although the adenine residue in the 3′ A2 ERE half site can serve as a hydrogen bond donor and acceptor, the guanine residue residing in a comparable position in the pS2 ERE can function only as a hydrogen bond acceptor. From previous crystallographic studies of the vitellogenin A2 ERE (36) and B1 ERE2 (38), one would predict that substitution of a guanine for an adenine in the 3′ ERE half site would not only affect the hydrogen bond with the substituted nucleotide but also require the modification of a localized hydrogen bond network formed between the ERE and the DBD. We have previously demonstrated that binding of the ER DBD and the full-length ER to the ERE induces conformational changes in DNA structure (25, 26, 28, 29, 33). We now provide evidence that the DBD-DNA interaction is a dynamic process involving conformational changes in both the receptor and DNA.
Our DNase I footprinting studies revealed that 1.3 pmol of DBD was required to occupy 50% of the 5′ A2 ERE half site. In contrast, 6.5 pmol of DBD was required to occupy 50% of the 5′ pS2 ERE half site. Despite the fact that these 5′ ERE half sites have identical nucleotide sequences, the relative affinity of the DBD is ∼5-fold lower for the 5′ pS2 ERE half site than for the 5′ A2 ERE half site (Fig. 3), suggesting that two adjacent consensus ERE half sites can act cooperatively to enhance DBD binding. When comparing the intact EREs, we found that the affinity of the DBD is more than sixfold greater for the two adjacent A2 ERE half sites than for the consensus and imperfect pS2 ERE half sites. We have previously demonstrated that the affinity of the intact ER is twofold lower for the pS2 ERE than for the A2 ERE (28). Thus, regions outside the DBD are important for enhancing binding of the intact receptor to the A2 ERE but may be even more important in enhancing binding of the receptor to imperfect ERE sequences.
We observed an apparent monomer-to-dimer transition as increasing concentrations of purified DBD were combined with the pS2 ERE. A similar monomer-to-dimer transition has been observed in experiments carried out with the ER DBD and the imperfect vitellogenin B1 ERE 2 (38), which differs from the consensus sequence by a single base pair in the 5′ half site (AGTCAnnnTGACC [50]). In contrast to the pS2 ERE and the B1 ERE2, we did not observe occupation of a single half site with the A2 ERE, implying that the DBD binds only as a dimer to the A2 ERE. Binding of the ER dimer to the A2 ERE has been a subject of substantial controversy. While NMR and crystal structure studies provide evidence for ER DBD dimer binding (36, 37), an antibody-based DNA binding assay (11) suggests that the ER may bind as a monomer to the A2 ERE. Taken together, our gel mobility, DNase I footprinting, and methylation interference assays examining the A2 and pS2 EREs in tandem provide compelling evidence that the DBD binds as a dimer to the A2 ERE and as a monomer and a dimer to the pS2 ERE.
Since we know that the DBD is a monomer in solution (27), dimerization must occur upon binding of the DBD to the ERE. Dimerization could be fostered by simultaneous binding of two DBD monomers or binding of one DBD monomer and the subsequent recruitment of a second DBD monomer. In either case, protein-protein and protein-DNA interactions would help stabilize binding of the DBD dimer and discourage dissociation of one of the monomers from the A2 or pS2 ERE. Monomer binding to the pS2 ERE could result from the inability of the DBD monomer bound to the consensus half site to recruit a second DBD monomer to the imperfect 3′ half site or more rapid dissociation of the DBD from the imperfect 3′ ERE half site.
From our combined experiments, it is possible to compare binding of the ER DBD and the full-length ER to the A2 and pS2 EREs. The ER DBD bound to the pS2 ERE as a monomer at low DBD concentrations and then as a dimer at higher DBD concentrations. These findings are similar to those of studies examining binding of the full-length ER to the vitellogenin B1 ERE2 (22). In contrast to these results, the full-length ER used in our experiments bound only as a dimer to the A2 and pS2 ERE, as indicated by the migration of the ER-ERE complexes in gel mobility shift assays (Fig. 6 and 7). The exclusive binding of the ER dimer to the pS2 ERE emphasizes that the dimerization domain present in the ligand binding domain (8) plays an important role in ER stabilization. Methylation interference assays demonstrated that guanine residues in both A2 ERE half sites were important for ER DBD binding. Since the exact same guanine residues are involved in binding of the full-length ER to the A2 ERE (17), the ER DBD and the full-length ER must bind to the A2 ERE in a very similar fashion.
Protease sensitivity assays demonstrated that there were distinct differences in the digestion patterns of the A2 and pS2 ERE-bound receptor. What is uncertain at this point is the conformational state of the individual ER monomers. We anticipate that the two ER monomers bound to the A2 ERE would have the same conformation. However, it is not known whether both of the ER monomers bound to the pS2 ERE have the same conformation. One might argue that the monomer bound to the consensus pS2 ERE half site would have the same conformation as the monomers bound to the consensus A2 ERE half sites but that the conformation of the monomer bound to the imperfect pS2 half site would be different. Alternatively, it is possible that binding of the ER monomer to the imperfect ERE half site would induce the formation of an altered dimerization interface, which would in turn cause conformational changes in the adjacent ER monomer. Our data favor this latter model since we do not see two superimposed digestion patterns, one for each ER monomer, after partial digestion of the pS2 ERE-bound receptor.
Antibody supershift experiments demonstrated that several antibodies directed at different ER epitopes enhanced ER-ERE binding. The ability of ER-specific antibodies to enhance ER-DNA complex formation has been previously reported by Fawell et al. (7), who suggested that this enhanced binding is due to stabilization of the ER dimer. P1A3 had the most dramatic effect on ER-ERE complex formation. It significantly enhanced ER binding to the A2 ERE, decreased ER binding to the pS2 ERE, and yet failed to supershift either ER-ERE complex. These results suggest that ER binding to antibody and binding to DNA are mutually exclusive events. The inability of P1A3 to supershift the ERE-bound ER was not unexpected, since this antibody is directed against the ER DBD and binding of the DBD to the ERE could presumably occlude the antibody epitope. The ability of P1A3 to enhance ER binding to the A2 ERE yet inhibit binding to the pS2 ERE was somewhat perplexing. However, a similar phenomenon in which an antibody directed against the vitamin D3 receptor DBD enhanced binding of receptor to the osteopontin response element and inhibited binding of receptor to the osteocalcin response element, but did not supershift either complex, has been reported (40). Staal et al. (40) proposed that the presence of the additional immunoglobulin G protein may have simply increased the association of the receptor for its cognate response element and thereby enhanced binding. However, we find that inclusion of additional nonspecific protein in our binding reactions did not affect ER-DNA complex formation (data not shown). It seems more probable that P1A3 enhanced A2 ERE binding by promoting ER dimerization and that binding of the ER dimer to the A2 ERE dissociated the antibody. The inability of the receptor to interact with the pS2 ERE in the presence of P1A3 may be attributed to more efficient binding of the ER to antibody than to the ERE or to an unfavorable presentation of the antibody-stabilized ER dimer to the pS2 ERE.
A number of studies have demonstrated that the activity of many ERE-containing promoters is cell type specific (3, 23, 24, 46, 47). It is generally thought that these tissue-specific effects are brought about by restricting the expression of required regulatory cofactors to target cells. A more versatile way of differentially regulating gene expression would be to provide the receptor with a large repertoire of functional surfaces that can be formed and serve as contact points for other cellular proteins. The presentation of these functional surfaces and the selection of ER-associated proteins, which is dictated by the unique ERE sequence, would provide tremendous regulatory versatility to a single cell harboring multiple estrogen-responsive genes.
We propose that the conformation of nuclear hormone receptors is subject to two ligands—hormone and DNA—and that binding of either ligand can induce changes in receptor conformation. The ability of hormone to induce conformational changes in nuclear receptor ligand binding domains has been demonstrated (2, 5, 14, 34, 49). Our studies with the ER complement those carried out with glucocorticoid and the vitamin D receptors and suggest that DNA-induced conformational changes in the DBD can be transmitted to other regions of the receptor (19, 40, 41). Taken together, these studies provide evidence that conformational changes induced by DNA binding may serve as a common mechanism for regulating transcription of hormone-responsive genes.
ACKNOWLEDGMENTS
This research was supported by NIH grant R29 HD31299 (to A.M.N.) and USMRMC grant DAMD17-94-J4228 (to G.L.G.). Jennifer Wood was supported by NIH Reproductive Training Grant PHS 2T32 HD 0728-19.
We thank David Shapiro, Dean Edwards, and Robin Fuchs-Young for providing ER-specific antibodies and Varsha Likhite for additional protease sensitivity experiments. We are also grateful to David Shapiro for helpful comments during preparation of the manuscript.
REFERENCES
- 1.Alber T. How GCN4 binds DNA. Curr Biol. 1993;3:182–184. doi: 10.1016/0960-9822(93)90268-s. [DOI] [PubMed] [Google Scholar]
- 2.Beekman J M, Allan G F, Tsai S Y, Tsai M-J, O’Malley B. Transcriptional activation by the estrogen receptor requires a conformational change in the ligand binding domain. Mol Endocrinol. 1993;7:1266–1274. doi: 10.1210/mend.7.10.8264659. [DOI] [PubMed] [Google Scholar]
- 3.Berry M, Metzger D, Chambon P. Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. EMBO J. 1990;9:2811–2818. doi: 10.1002/j.1460-2075.1990.tb07469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaustein J. Estrogen receptor immunoreactivity in rat brain: rapid effects of estradiol injection. Endocrinology. 1993;132:1218–1224. doi: 10.1210/endo.132.3.7679973. [DOI] [PubMed] [Google Scholar]
- 5.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structural of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 6.Chang T-C, Nardulli A M, Lew D, Shapiro D J. The role of estrogen response elements in expression of the Xenopus laevis vitellogenin B1 gene. Mol Endocrinol. 1992;6:346–354. doi: 10.1210/mend.6.3.1584211. [DOI] [PubMed] [Google Scholar]
- 7.Fawell S, White R, Hoare S, Sydenham M, Page M, Parker M. Inhibition of estrogen receptor-DNA binding by the “pure” antiestrogen ICI 164,384 appears to be mediated by impaired receptor dimerization. Proc Natl Acad Sci USA. 1990;17:6883–6887. doi: 10.1073/pnas.87.17.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fawell S E, Lees J A, White R, Parker M G. Characterization and colocalization of steroid binding and dimerization activities in the mouse estrogen receptor. Cell. 1990;60:953–962. doi: 10.1016/0092-8674(90)90343-d. [DOI] [PubMed] [Google Scholar]
- 9.Frankel A D, Kim P S. Modular structure of transcription factors: implications for gene regulation. Cell. 1991;65:717–719. doi: 10.1016/0092-8674(91)90378-c. [DOI] [PubMed] [Google Scholar]
- 10.Fujita T, Nolan G P, Ghosh S, Baltimore D. Independent modes of transcriptional activation by the p50 and p65 subunits of NF-KB. Genes Dev. 1992;6:775–787. doi: 10.1101/gad.6.5.775. [DOI] [PubMed] [Google Scholar]
- 11.Furlow J D, Murdoch F E, Gorski J. High affinity binding of the estrogen receptor to a DNA response element does not require homodimer formation or estrogen. J Biol Chem. 1993;268:12519–12525. [PubMed] [Google Scholar]
- 12.Green S, Chambon P. Oestradiol induction of a glucocorticoid-responsive gene by a chimaeric receptor. Nature. 1987;325:75–78. doi: 10.1038/325075a0. [DOI] [PubMed] [Google Scholar]
- 13.Greene G, Sobel N, King W, Jensen E. Immunochemical studies of estrogen receptors. J Steroid Biochem. 1984;20:51–56. doi: 10.1016/0022-4731(84)90188-2. [DOI] [PubMed] [Google Scholar]
- 14.Hansen J C, Gorski J. Conformational transitions of the estrogen receptor monomer. J Biol Chem. 1986;261:13990–13996. [PubMed] [Google Scholar]
- 15.Hard T, Kellenbach E, Boelens R, Naler B A, Dahlman K, Freedman L P, Carlstedt-Duke J, Yamamoto K R, Gustafsson J-A, Kaptein R. Solution structure of the glucocorticoid receptor DNA-binding domain. Science. 1990;249:157–160. doi: 10.1126/science.2115209. [DOI] [PubMed] [Google Scholar]
- 16.Klein-Hitpass L, Ryffel G U, Heitlinger E, Cato A C B. A 13 bp palindrome is a functional estrogen responsive element and interacts specifically with estrogen receptor. Nucleic Acids Res. 1988;16:647–663. doi: 10.1093/nar/16.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein-Hitpass L, Tsai S Y, Greene G L, Clark J H, Tsai M-J, O’Malley B W. Specific binding of estrogen receptor to the estrogen response element. Mol Cell Biol. 1989;9:43–49. doi: 10.1128/mcb.9.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuiper G G J M, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson J-A. Cloning of a novel estrogen receptor expressed in rat prostate and ovary. Proc Natl Acad Sci USA. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefstin J A, Thomas J R, Yamamoto K R. Influence of a steroid receptor DNA-binding domain on transcriptional regulatory functions. Genes Dev. 1994;8:2842–2856. doi: 10.1101/gad.8.23.2842. [DOI] [PubMed] [Google Scholar]
- 20.Littlewood T D, Hancock D C, Danielian P S, Parker M G, Evan G I. A modified oestrogen receptor ligand binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luisi B F, Xu W X, Otwinowski Z, Freedman L P, Yamamoto K R, Sigler P B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 22.Martinez E, Wahli W. Cooperative binding of estrogen receptor to imperfect estrogen-responsive DNA elements correlates with their synergistic hormone-dependent enhancer activity. EMBO J. 1989;8:3781–3791. doi: 10.1002/j.1460-2075.1989.tb08555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metzger D, Ali S, Bornert J-M, Chambon P. Characterization of the amino-terminal transcriptional activation function of the human estrogen receptor in animal and yeast cells. J Biol Chem. 1995;270:9535–9542. doi: 10.1074/jbc.270.16.9535. [DOI] [PubMed] [Google Scholar]
- 24.Montano M M, Ekena K, Krueger K D, Keller A L, Katzenellenbogen B S. Human estrogen receptor ligand activity inversion mutants: receptors that interpret antiestrogens as estrogens and estrogens as antiestrogens and discriminate among different antiestrogens. Mol Endocrinol. 1996;10:230–242. doi: 10.1210/mend.10.3.8833652. [DOI] [PubMed] [Google Scholar]
- 25.Nardulli A M, Greene G L, Shapiro D J. Human estrogen receptor bound to an estrogen response element bends DNA. Mol Endocrinol. 1993;7:331–340. doi: 10.1210/mend.7.3.8483477. [DOI] [PubMed] [Google Scholar]
- 26.Nardulli A M, Grobner C, Cotter D. Estrogen receptor-induced DNA bending: orientation of the bend and replacement of an estrogen response element with an intrinsic DNA bending sequence. Mol Endocrinol. 1995;9:1064–1076. doi: 10.1210/mend.9.8.7476980. [DOI] [PubMed] [Google Scholar]
- 27.Nardulli A M, Lew D, Erijman L, Shapiro D J. Purified estrogen receptor DNA binding domain expressed in Escherichia coli activates transcription of an estrogen-responsive promoter in cultured cells. J Biol Chem. 1991;266:24070–24076. [PubMed] [Google Scholar]
- 28.Nardulli A M, Romine L E, Carpo C, Greene G L, Rainish B. Estrogen receptor affinity and location of consensus and imperfect estrogen response elements influence transcription activation of simplified promoters. Mol Endocrinol. 1996;10:694–704. doi: 10.1210/mend.10.6.8776729. [DOI] [PubMed] [Google Scholar]
- 29.Nardulli A M, Shapiro D J. Binding of the estrogen receptor DNA-binding domain to the estrogen response element induces DNA bending. Mol Cell Biol. 1992;12:2037–2042. doi: 10.1128/mcb.12.5.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunez A-M, Berry M, Imler J-L, Chambon P. The 5′ flanking region of the pS2 gene contains a complex enhancer region responsive to oestrogens, epidermal growth factor, a tumour promoter (TPA), the c-Ha-ras oncoprotein and the c-jun protein. EMBO J. 1989;8:823–829. doi: 10.1002/j.1460-2075.1989.tb03443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pabo C O, Sauer R T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 32.Ponglikitmongkol M, Green S, Chambon P. Genomic organization of the human oestrogen receptor gene. EMBO J. 1988;7:3385–3388. doi: 10.1002/j.1460-2075.1988.tb03211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potthoff S J, Romine L E, Nardulli A M. Effects of wild type and mutant estrogen receptors on DNA flexibility, DNA bending, and transcription activation. Mol Endocrinol. 1996;10:1095–1106. doi: 10.1210/mend.10.9.8885244. [DOI] [PubMed] [Google Scholar]
- 34.Renaud J-P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR- gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 35.Schreiber E, Matthias P, Müller M, Schaffner W. Identification of a novel lymphoid specific octamer binding protein (OTF-2B) by proteolytic clipping bandshift assay (PCBA) EMBO J. 1988;7:4221–4229. doi: 10.1002/j.1460-2075.1988.tb03319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwabe J W R, Chapman L, Finch J T, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 37.Schwabe J W R, Chapman L, Finch J T, Rhodes D, Neuhaus D. DNA recognition by the oestrogen receptor: From solution to the crystal. Structure. 1993;1:187–204. doi: 10.1016/0969-2126(93)90020-h. [DOI] [PubMed] [Google Scholar]
- 38.Schwabe J W R, Chapman L, Rhodes D. The oestrogen receptor recognizes an imperfectly palindromic response element through an alternative side-chain conformation. Structure. 1995;3:201–213. doi: 10.1016/s0969-2126(01)00150-2. [DOI] [PubMed] [Google Scholar]
- 39.Spolar R S, Record M T. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 40.Staal A, van Wijnen A J, Birkenhäger J C, Pols H A P, Prahl J, DeLuca H, Gaub M-P, Lian J B, Stein G S, van Leeuwen J P T M, Stein J L. Distinct conformations of vitamin D receptor/retinoid X receptor-α heterodimers are specified by dinucleotide differences in the vitamin D-responsive elements of the osteocalcin and osteopontin genes. Mol Endocrinol. 1996;10:1444–1456. doi: 10.1210/mend.10.11.8923469. [DOI] [PubMed] [Google Scholar]
- 41.Starr D B, Matsui W, Thomas J R, Yamamoto K R. Intracellular receptors use a common mechanism to interpret signaling information at response elements. Genes Dev. 1996;10:1271–1283. doi: 10.1101/gad.10.10.1271. [DOI] [PubMed] [Google Scholar]
- 42.Steitz T A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990;23:205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- 43.Talanian R V, McKnight C J, Kim P S. Sequence-specific DNA binding by a short peptide dimer. Science. 1990;249:769–771. doi: 10.1126/science.2389142. [DOI] [PubMed] [Google Scholar]
- 44.Tan S, Richmond T J. DNA binding-induced conformational change of the yeast transcriptional activator PRTF. Cell. 1990;62:367–377. doi: 10.1016/0092-8674(90)90373-m. [DOI] [PubMed] [Google Scholar]
- 45.Tasset D, Tora L, Fromental C, Scheer E, Chambon P. Distinct classes of transcriptional activating domains function by different mechanisms. Cell. 1990;62:1177–1187. doi: 10.1016/0092-8674(90)90394-t. [DOI] [PubMed] [Google Scholar]
- 46.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 47.Tzukerman M T, Esty A, Santiso-Mere D, Danielian P, Parker M G, Stein R B, Pike J W, McDonnell D P. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol. 1994;8:21–30. doi: 10.1210/mend.8.1.8152428. [DOI] [PubMed] [Google Scholar]
- 48.von Hippel P H. Protein-DNA recognition: new perspectives and underlying themes. Science. 1994;263:769–770. doi: 10.1126/science.8303292. [DOI] [PubMed] [Google Scholar]
- 49.Wagner R, Apriletti J, McGrath M, West B, Baxter J, Fletterick R. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 50.Walker P, Germond J-E, Brown-Luedi M, Givel F, Wahli W. Sequence homologies in the region preceding the transcription initiation site of the liver estrogen-responsive vitellogenin and apo-VLDLII genes. Nucleic Acids Res. 1984;12:8611–8626. doi: 10.1093/nar/12.22.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webster N J G, Green S, Jin J R, Chambon P. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell. 1988;54:199–207. doi: 10.1016/0092-8674(88)90552-1. [DOI] [PubMed] [Google Scholar]