The CdII atom in the title complex [Cd(NO3)2(2AB)4] (2AB is 2-aminobenzaxole; C7H6N2O) has a distorted octahedral coordination environment. In the crystal structure, several N—H⋯O interactions lead to the formation of layers parallel to (001).
Keywords: crystal structure, molecular structure, cadmium complex, 2-aminobenzoxazole, octahedral coordination
Abstract
A coordination complex of cadmium nitrate [Cd(NO3)2] with 2-aminobenzaxole (2AB; C7H6N2O), namely, tetrakis(2-aminobenzoxazole-κN1)bis(nitrato-κO)cadmium(II), [Cd(NO3)2(2AB)4], has been synthesized from ethanol solutions of Cd(NO3)2·H2O and 2AB. The asymmetric unit comprises half a molecule of [Cd(NO3)2(2AB)4], with the CdII atom positioned on a twofold rotation axis. In the completed molecular complex, four 2AB ligands and two nitrate anions each coordinate monodentately to the CdII atom, leading to a distorted octahedral coordination environment. The crystal structure of [Cd(NO3)2(2AB)4] exhibits several N—H⋯O interactions, resulting in the formation of a layered assembly parallel to (001). Hishfeld surface analysis was used to quantify the intermolecular interactions.
1. Chemical context
Benzoxazole is a heterocyclic aromatic compound consisting of a benzene ring fused to an oxazole ring. It has a strong and unpleasant fishy odour, just like pyridine (Katritzky & Pozharskii, 2000 ▸; Clayden et al., 2001 ▸). Many benzoxazole-based compounds are valued in medicinal and biological research because of their numerous biological activities (Potashman et al., 2007 ▸; Šlachtová. & Brulíková, 2018 ▸; Razzoqova et al., 2022 ▸, 2024 ▸), including antimicrobial (Erol et al., 2022 ▸), antitumor (Imaizumi et al., 2020 ▸), anti-inflammatory (Parlapalli & Manda, 2017 ▸), analgesic (Ali et al., 2022 ▸; Sattar et al., 2020 ▸), antitubercular (Šlachtová & Brulíková, 2018 ▸), herbicidal (Sangi et al., 2019 ▸), and fungicidal properties (Fan et al., 2022 ▸).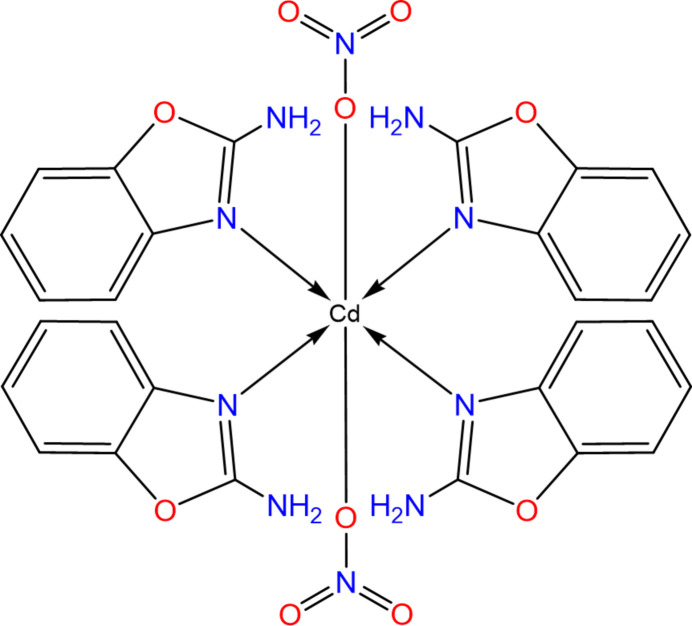 At the same time, 2-aminobenzoxazole (2AB) and its derivatives have potent antibacterial and anticancer properties (Paramashivappa et al., 2003 ▸; Khajondetchairit et al., 2017 ▸; Ouyang et al., 2012 ▸). One notable derivative of 2AB is 2-amino-5-chlorobenzoxazole, which has demonstrated muscle relaxant effects and is used as an antispasmodic and uricosuric agent in therapeutic applications (Lynch, 2004 ▸).
At the same time, 2-aminobenzoxazole (2AB) and its derivatives have potent antibacterial and anticancer properties (Paramashivappa et al., 2003 ▸; Khajondetchairit et al., 2017 ▸; Ouyang et al., 2012 ▸). One notable derivative of 2AB is 2-amino-5-chlorobenzoxazole, which has demonstrated muscle relaxant effects and is used as an antispasmodic and uricosuric agent in therapeutic applications (Lynch, 2004 ▸).
In the context given above, we present here the synthesis, crystal structure determination and Hirshfeld surface analysis of a coordination complex of 2AB with cadmium nitrate, [Cd(NO3)2(2AB)4].
2. Structural commentary
In the asymmetric unit of [Cd(NO3)2(2AB)4], which consists of half of a complex molecule, the CdII atom is positioned on a twofold rotation axis (multiplicity 4, Wyckoff letter e). In the completed molecule, the CdII atom coordinates by four 2AB ligands and two nitrate anions, resulting in a distorted octahedral N4O2 coordination set (Fig. 1 ▸). The four 2AB ligands occupy the equatorial positions and are coordinated monodentately through their aromatic nitrogen donor atoms with Cd—N bond lengths of 2.314 (3) and 2.325 (3) Å. The two axially positioned nitrato ligands are also coordinated in a monodentate fashion with a relatively long Cd—O bond length of 2.418 (3) Å. The dihedral angle formed between the two opposite 2-aminobenzaxazole ligands (labelled in Fig. 1 ▸) is 84.85 (17)°. The molecular conformation is stabilized by intramolecular N—H⋯O hydrogen-bonding interactions involving the coordinated oxygen atom O1 and the non-coordinated oxygen atom O2 (entries #1 and #3 in Table 1 ▸).
Figure 1.
The molecular structure of [Cd(NO3)2(2AB)4] with displacement ellipsoids drawn at the 30% probability level; non-labelled atoms are generated by symmetry code −x + 1, y, −z +  . Intramolecular hydrogen bonds are indicated by dashed blue lines.
. Intramolecular hydrogen bonds are indicated by dashed blue lines.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N4—H4A⋯O1i | 0.86 | 2.26 | 2.971 (5) | 140 |
| N4—H4B⋯O2ii | 0.86 | 2.28 | 2.822 (6) | 121 |
| N2—H2A⋯O2i | 0.86 | 2.11 | 2.899 (7) | 152 |
| N2—H2B⋯O3iii | 0.86 | 2.33 | 2.953 (6) | 129 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii) 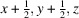 .
.
3. Supramolecular features
In the crystal structure of [Cd(NO3)2(2AB)4], intermolecular N—H⋯O hydrogen bonds involving the non-coordinated O atoms O2 and O3 (entries #2 and #4 in Table 1 ▸) lead to the formation of sheets extending parallel to (001), as shown in Fig. 2 ▸.
Figure 2.
Visualization of the molecular packing in [Cd(NO3)2(2AB)4] in a view along [010]. Intermolecular N—H⋯O interactions are shows as light-blue dashed lines.
4. Hirshfeld Surface Analysis
Hirshfeld surface (HS) analysis (Spackman & Jayatilaka, 2009 ▸) was performed and two-dimensional fingerprint plots (Spackman & McKinnon, 2002 ▸) were generated using CrystalExplorer (Spackman et al., 2021 ▸) to quantify the intermolecular interactions. HS and fingerprint plot analysis conducted for [Cd(NO3)2(2AB)4] are graphically displayed in Fig. 3 ▸. The red spots on the HS area of [Cd(NO3)2(2AB)4] confirm the close intermolecular N—H⋯O contacts (related to entries #2 and #4 in Table 1 ▸) between adjacent molecules. The two-dimensional fingerprint plots and their relative contributions revealed that H⋯H, O⋯H, C⋯H, C⋯O, O⋯O and N⋯H interactions are the main interactions to the HS area. Specifically, the fingerprint plots reveal the presence of close N—H⋯O contacts in form of two spikes observed near (di + de) ≃ 2.3 Å and C—H contacts as two wings near (di + de) ≃ 2. 8 Å (Fig. 3 ▸).
Figure 3.
View of HS and two-dimensional fingerprint plots of [Cd(NO3)2(2AB)4].
5. Database survey
A survey of the Cambridge Structural Database (CSD, Version 5.46, November 2024; Groom et al., 2016 ▸) revealed 17 crystal structures of 2-aminobenzoxazole derivatives. Among these, only two structures involve coordination compounds with zinc (QALXIL; Decken & Gossage, 2005 ▸) and cadmium (DIWPIM; Razzoqova et al., 2023 ▸). In the zinc complex, the central metal atom coordinates two benzoxazolamine ligands through the aromatic nitrogen atom and two chloro ligands in a distorted tetrahedral coordination environment. In the crystal structure of DIWPIM, which corresponds to [Cd(2AB)2(CH3COO)2], the CdII atom coordinates by two 2AB ligands and two acetato ligands in a monodentate and bidentate fashion, respectively, forming a distorted octahedral N2O4 coordination set.
6. Synthesis and crystallization
Cd(NO3)2·H2O (0.308 g, 1 mmol) and 2AB (0.268 g, 2 mmol) were dissolved separately in ethanol (5 ml), mixed together and stirred for 2 h. The obtained colourless solution was filtered and left for crystallization. Single crystals of the complex [Cd(NO3)2(2AB)4] suitable for X-ray analysis were obtained by slow evaporation of the solution over a period of 7 d.
7. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. Hydrogen atoms were treated in a riding model with Uiso(H) = 1.2Ueq(C,N).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | [Cd(NO3)2(C7H6N2O)4] |
| M r | 772.97 |
| Crystal system, space group | Monoclinic, C2/c |
| Temperature (K) | 293 |
| a, b, c (Å) | 15.9012 (3), 11.0897 (2), 18.9475 (5) |
| β (°) | 109.182 (3) |
| V (Å3) | 3155.70 (13) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 6.19 |
| Crystal size (mm) | 0.10 × 0.08 × 0.06 |
| Data collection | |
| Diffractometer | XtaLAB Synergy, Single source at home/near, HyPix3000 |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2020 ▸) |
| Tmin, Tmax | 0.016, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 12616, 3014, 2324 |
| R int | 0.084 |
| (sin θ/λ)max (Å−1) | 0.614 |
| Refinement | |
| R[F2 > 2σ(F2)], wR(F2), S | 0.049, 0.128, 1.01 |
| No. of reflections | 3014 |
| No. of parameters | 223 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.56, −0.95 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989025004049/wm5756sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989025004049/wm5756Isup2.hkl
CCDC reference: 2448896
Additional supporting information: crystallographic information; 3D view; checkCIF report
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
BT would like to acknowledge the CSIR–TWAS fellowship and the FAIRE programme provided by the Cambridge Crystallographic Data Centre (CCDC) for the use of the Cambridge Structural Database (CSD) and associated software.
supplementary crystallographic information
Tetrakis(2-aminobenzoxazole-κN1)bis(nitrato-κO)cadmium(II) . Crystal data
| [Cd(NO3)2(C7H6N2O)4] | F(000) = 1560 |
| Mr = 772.97 | Dx = 1.627 Mg m−3 |
| Monoclinic, C2/c | Cu Kα radiation, λ = 1.54184 Å |
| a = 15.9012 (3) Å | Cell parameters from 4500 reflections |
| b = 11.0897 (2) Å | θ = 4.9–70.8° |
| c = 18.9475 (5) Å | µ = 6.19 mm−1 |
| β = 109.182 (3)° | T = 293 K |
| V = 3155.70 (13) Å3 | Block, colourless |
| Z = 4 | 0.10 × 0.08 × 0.06 mm |
Tetrakis(2-aminobenzoxazole-κN1)bis(nitrato-κO)cadmium(II) . Data collection
| XtaLAB Synergy, Single source at home/near, HyPix3000 diffractometer | 3014 independent reflections |
| Radiation source: micro-focus sealed X-ray tube, PhotonJet (Cu) X-ray Source | 2324 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.084 |
| Detector resolution: 10.0000 pixels mm-1 | θmax = 71.1°, θmin = 4.9° |
| ω scans | h = −19→19 |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2020) | k = −13→13 |
| Tmin = 0.016, Tmax = 1.000 | l = −22→23 |
| 12616 measured reflections |
Tetrakis(2-aminobenzoxazole-κN1)bis(nitrato-κO)cadmium(II) . Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.049 | w = 1/[σ2(Fo2) + (0.0631P)2] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.128 | (Δ/σ)max < 0.001 |
| S = 1.01 | Δρmax = 0.56 e Å−3 |
| 3014 reflections | Δρmin = −0.95 e Å−3 |
| 223 parameters | Extinction correction: SHELXL (Sheldrick, 2015b), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.00041 (6) |
Tetrakis(2-aminobenzoxazole-κN1)bis(nitrato-κO)cadmium(II) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Tetrakis(2-aminobenzoxazole-κN1)bis(nitrato-κO)cadmium(II) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cd1 | 0.500000 | 0.24505 (3) | 0.750000 | 0.04211 (19) | |
| O5 | 0.5198 (2) | −0.0974 (3) | 0.6354 (2) | 0.0633 (9) | |
| O4 | 0.5320 (2) | 0.5949 (3) | 0.8680 (2) | 0.0650 (9) | |
| O1 | 0.3406 (2) | 0.2156 (3) | 0.7009 (2) | 0.0639 (9) | |
| N1 | 0.5181 (2) | 0.3989 (3) | 0.8371 (2) | 0.0461 (8) | |
| N3 | 0.5049 (2) | 0.0945 (3) | 0.6668 (2) | 0.0470 (8) | |
| N5 | 0.2779 (2) | 0.2434 (4) | 0.7254 (3) | 0.0572 (11) | |
| N4 | 0.6112 (2) | −0.0366 (4) | 0.7492 (2) | 0.0625 (11) | |
| H4A | 0.629684 | 0.016170 | 0.784177 | 0.075* | |
| H4B | 0.634011 | −0.107721 | 0.755194 | 0.075* | |
| O2 | 0.2525 (3) | 0.3475 (4) | 0.7173 (3) | 0.1036 (16) | |
| N2 | 0.6252 (3) | 0.5215 (4) | 0.8094 (3) | 0.0752 (13) | |
| H2A | 0.645052 | 0.464034 | 0.788780 | 0.090* | |
| H2B | 0.647782 | 0.592541 | 0.812309 | 0.090* | |
| O3 | 0.2472 (3) | 0.1686 (5) | 0.7563 (3) | 0.1067 (16) | |
| C5 | 0.3927 (3) | 0.3589 (5) | 0.8882 (3) | 0.0590 (12) | |
| H5 | 0.386358 | 0.277102 | 0.876857 | 0.071* | |
| C6 | 0.4556 (3) | 0.4272 (4) | 0.8722 (2) | 0.0483 (10) | |
| C13 | 0.4434 (3) | 0.0736 (4) | 0.5961 (3) | 0.0487 (10) | |
| C7 | 0.5601 (3) | 0.5007 (4) | 0.8366 (3) | 0.0527 (11) | |
| C12 | 0.3822 (3) | 0.1470 (5) | 0.5465 (3) | 0.0560 (11) | |
| H12 | 0.374376 | 0.226672 | 0.558271 | 0.067* | |
| C1 | 0.4640 (3) | 0.5490 (4) | 0.8903 (3) | 0.0591 (12) | |
| C2 | 0.4121 (4) | 0.6093 (5) | 0.9230 (3) | 0.0768 (16) | |
| H2 | 0.418750 | 0.691342 | 0.933603 | 0.092* | |
| C11 | 0.3324 (3) | 0.0975 (6) | 0.4779 (3) | 0.0754 (16) | |
| H11 | 0.291093 | 0.145209 | 0.442908 | 0.091* | |
| C14 | 0.5477 (3) | −0.0084 (4) | 0.6863 (3) | 0.0501 (11) | |
| C8 | 0.4530 (3) | −0.0457 (4) | 0.5772 (3) | 0.0595 (12) | |
| C4 | 0.3384 (4) | 0.4174 (6) | 0.9221 (3) | 0.0793 (17) | |
| H4 | 0.294517 | 0.373897 | 0.933595 | 0.095* | |
| C3 | 0.3487 (4) | 0.5396 (7) | 0.9391 (4) | 0.087 (2) | |
| H3 | 0.311723 | 0.575692 | 0.962182 | 0.105* | |
| C10 | 0.3436 (4) | −0.0227 (7) | 0.4611 (4) | 0.086 (2) | |
| H10 | 0.308806 | −0.053870 | 0.415260 | 0.103* | |
| C9 | 0.4051 (4) | −0.0967 (6) | 0.5107 (3) | 0.0831 (18) | |
| H9 | 0.413245 | −0.176568 | 0.499456 | 0.100* |
Tetrakis(2-aminobenzoxazole-κN1)bis(nitrato-κO)cadmium(II) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cd1 | 0.0425 (2) | 0.0410 (3) | 0.0463 (3) | 0.000 | 0.01946 (17) | 0.000 |
| O5 | 0.068 (2) | 0.0510 (17) | 0.074 (2) | 0.0032 (16) | 0.0276 (18) | −0.0150 (17) |
| O4 | 0.073 (2) | 0.0507 (18) | 0.074 (2) | −0.0063 (16) | 0.0271 (18) | −0.0097 (17) |
| O1 | 0.0380 (16) | 0.081 (2) | 0.075 (2) | 0.0023 (15) | 0.0215 (16) | −0.0023 (18) |
| N1 | 0.0500 (19) | 0.0458 (18) | 0.046 (2) | −0.0011 (16) | 0.0208 (16) | −0.0034 (16) |
| N3 | 0.0495 (18) | 0.0491 (19) | 0.047 (2) | 0.0014 (16) | 0.0214 (16) | −0.0052 (16) |
| N5 | 0.0346 (17) | 0.077 (3) | 0.060 (2) | 0.0003 (18) | 0.0162 (17) | 0.000 (2) |
| N4 | 0.059 (2) | 0.053 (2) | 0.075 (3) | 0.0158 (18) | 0.022 (2) | 0.002 (2) |
| O2 | 0.100 (3) | 0.096 (3) | 0.126 (4) | 0.050 (3) | 0.053 (3) | 0.018 (3) |
| N2 | 0.068 (3) | 0.073 (3) | 0.094 (4) | −0.025 (2) | 0.039 (3) | −0.017 (3) |
| O3 | 0.107 (3) | 0.113 (4) | 0.126 (4) | −0.044 (3) | 0.074 (3) | −0.008 (3) |
| C5 | 0.063 (3) | 0.066 (3) | 0.053 (3) | 0.001 (2) | 0.026 (2) | 0.002 (2) |
| C6 | 0.049 (2) | 0.056 (2) | 0.041 (2) | 0.006 (2) | 0.0149 (18) | −0.0022 (19) |
| C13 | 0.046 (2) | 0.054 (2) | 0.049 (3) | −0.0074 (19) | 0.0186 (19) | −0.003 (2) |
| C7 | 0.053 (2) | 0.052 (2) | 0.051 (3) | −0.007 (2) | 0.014 (2) | −0.007 (2) |
| C12 | 0.058 (3) | 0.067 (3) | 0.049 (3) | −0.005 (2) | 0.025 (2) | −0.001 (2) |
| C1 | 0.062 (3) | 0.055 (3) | 0.058 (3) | 0.008 (2) | 0.018 (2) | −0.001 (2) |
| C2 | 0.084 (4) | 0.073 (4) | 0.077 (4) | 0.017 (3) | 0.032 (3) | −0.009 (3) |
| C11 | 0.061 (3) | 0.106 (5) | 0.060 (3) | −0.008 (3) | 0.019 (3) | 0.003 (3) |
| C14 | 0.046 (2) | 0.048 (2) | 0.062 (3) | 0.0033 (18) | 0.025 (2) | −0.004 (2) |
| C8 | 0.062 (3) | 0.058 (3) | 0.067 (3) | −0.007 (2) | 0.031 (2) | −0.015 (2) |
| C4 | 0.075 (3) | 0.100 (5) | 0.071 (4) | −0.005 (3) | 0.035 (3) | 0.000 (3) |
| C3 | 0.083 (4) | 0.103 (5) | 0.084 (4) | 0.027 (4) | 0.038 (3) | −0.017 (4) |
| C10 | 0.073 (4) | 0.114 (5) | 0.067 (4) | −0.019 (4) | 0.019 (3) | −0.033 (4) |
| C9 | 0.079 (4) | 0.084 (4) | 0.086 (5) | −0.016 (3) | 0.027 (3) | −0.035 (3) |
Tetrakis(2-aminobenzoxazole-κN1)bis(nitrato-κO)cadmium(II) . Geometric parameters (Å, º)
| Cd1—O1i | 2.418 (3) | N2—C7 | 1.319 (6) |
| Cd1—O1 | 2.418 (3) | C5—H5 | 0.9300 |
| Cd1—N1i | 2.325 (3) | C5—C6 | 1.365 (6) |
| Cd1—N1 | 2.325 (3) | C5—C4 | 1.396 (7) |
| Cd1—N3 | 2.314 (3) | C6—C1 | 1.390 (6) |
| Cd1—N3i | 2.314 (3) | C13—C12 | 1.375 (6) |
| O5—C14 | 1.350 (5) | C13—C8 | 1.392 (6) |
| O5—C8 | 1.380 (6) | C12—H12 | 0.9300 |
| O4—C7 | 1.349 (5) | C12—C11 | 1.392 (7) |
| O4—C1 | 1.381 (6) | C1—C2 | 1.358 (7) |
| O1—N5 | 1.269 (5) | C2—H2 | 0.9300 |
| N1—C6 | 1.401 (6) | C2—C3 | 1.383 (8) |
| N1—C7 | 1.314 (5) | C11—H11 | 0.9300 |
| N3—C13 | 1.395 (6) | C11—C10 | 1.396 (9) |
| N3—C14 | 1.318 (5) | C8—C9 | 1.363 (7) |
| N5—O2 | 1.217 (5) | C4—H4 | 0.9300 |
| N5—O3 | 1.206 (6) | C4—C3 | 1.391 (8) |
| N4—H4A | 0.8600 | C3—H3 | 0.9300 |
| N4—H4B | 0.8600 | C10—H10 | 0.9300 |
| N4—C14 | 1.322 (6) | C10—C9 | 1.381 (9) |
| N2—H2A | 0.8600 | C9—H9 | 0.9300 |
| N2—H2B | 0.8600 | ||
| O1—Cd1—O1i | 164.46 (17) | C1—C6—N1 | 108.1 (4) |
| N1—Cd1—O1i | 87.51 (12) | C12—C13—N3 | 132.4 (4) |
| N1—Cd1—O1 | 104.00 (12) | C12—C13—C8 | 119.9 (4) |
| N1i—Cd1—O1i | 104.00 (12) | C8—C13—N3 | 107.7 (4) |
| N1i—Cd1—O1 | 87.51 (12) | N1—C7—O4 | 114.8 (4) |
| N1i—Cd1—N1 | 85.58 (18) | N1—C7—N2 | 128.3 (4) |
| N3—Cd1—O1 | 84.62 (12) | N2—C7—O4 | 116.9 (4) |
| N3—Cd1—O1i | 84.18 (13) | C13—C12—H12 | 121.2 |
| N3i—Cd1—O1 | 84.18 (13) | C13—C12—C11 | 117.6 (5) |
| N3i—Cd1—O1i | 84.63 (12) | C11—C12—H12 | 121.2 |
| N3—Cd1—N1 | 171.33 (11) | O4—C1—C6 | 107.7 (4) |
| N3i—Cd1—N1 | 94.01 (14) | C2—C1—O4 | 127.7 (5) |
| N3i—Cd1—N1i | 171.33 (11) | C2—C1—C6 | 124.6 (5) |
| N3—Cd1—N1i | 94.01 (14) | C1—C2—H2 | 122.5 |
| N3i—Cd1—N3 | 87.70 (18) | C1—C2—C3 | 115.0 (6) |
| C14—O5—C8 | 104.7 (4) | C3—C2—H2 | 122.5 |
| C7—O4—C1 | 104.9 (4) | C12—C11—H11 | 119.6 |
| N5—O1—Cd1 | 132.6 (3) | C12—C11—C10 | 120.8 (6) |
| C6—N1—Cd1 | 124.1 (3) | C10—C11—H11 | 119.6 |
| C7—N1—Cd1 | 124.8 (3) | N3—C14—O5 | 114.5 (4) |
| C7—N1—C6 | 104.6 (4) | N3—C14—N4 | 129.0 (4) |
| C13—N3—Cd1 | 127.1 (3) | N4—C14—O5 | 116.5 (4) |
| C14—N3—Cd1 | 124.3 (3) | O5—C8—C13 | 108.1 (4) |
| C14—N3—C13 | 105.1 (4) | C9—C8—O5 | 128.0 (5) |
| O2—N5—O1 | 116.9 (5) | C9—C8—C13 | 123.9 (5) |
| O3—N5—O1 | 120.2 (5) | C5—C4—H4 | 119.5 |
| O3—N5—O2 | 122.9 (5) | C3—C4—C5 | 121.0 (6) |
| H4A—N4—H4B | 120.0 | C3—C4—H4 | 119.5 |
| C14—N4—H4A | 120.0 | C2—C3—C4 | 122.2 (6) |
| C14—N4—H4B | 120.0 | C2—C3—H3 | 118.9 |
| H2A—N2—H2B | 120.0 | C4—C3—H3 | 118.9 |
| C7—N2—H2A | 120.0 | C11—C10—H10 | 119.1 |
| C7—N2—H2B | 120.0 | C9—C10—C11 | 121.8 (5) |
| C6—C5—H5 | 121.5 | C9—C10—H10 | 119.1 |
| C6—C5—C4 | 117.0 (5) | C8—C9—C10 | 116.0 (6) |
| C4—C5—H5 | 121.5 | C8—C9—H9 | 122.0 |
| C5—C6—N1 | 131.8 (4) | C10—C9—H9 | 122.0 |
| C5—C6—C1 | 120.2 (5) | ||
| Cd1—O1—N5—O2 | 79.2 (6) | C13—N3—C14—O5 | −0.8 (5) |
| Cd1—O1—N5—O3 | −99.7 (5) | C13—N3—C14—N4 | −178.2 (5) |
| Cd1—N1—C6—C5 | 26.7 (6) | C13—C12—C11—C10 | 0.9 (8) |
| Cd1—N1—C6—C1 | −151.9 (3) | C13—C8—C9—C10 | −0.6 (9) |
| Cd1—N1—C7—O4 | 152.3 (3) | C7—O4—C1—C6 | 1.0 (5) |
| Cd1—N1—C7—N2 | −27.5 (7) | C7—O4—C1—C2 | −177.8 (5) |
| Cd1—N3—C13—C12 | 22.8 (7) | C7—N1—C6—C5 | 179.3 (5) |
| Cd1—N3—C13—C8 | −158.9 (3) | C7—N1—C6—C1 | 0.7 (5) |
| Cd1—N3—C14—O5 | 159.4 (3) | C12—C13—C8—O5 | 178.4 (4) |
| Cd1—N3—C14—N4 | −18.0 (7) | C12—C13—C8—C9 | 0.5 (8) |
| O5—C8—C9—C10 | −178.0 (5) | C12—C11—C10—C9 | −1.1 (9) |
| O4—C1—C2—C3 | 179.8 (5) | C1—O4—C7—N1 | −0.6 (5) |
| N1—C6—C1—O4 | −1.1 (5) | C1—O4—C7—N2 | 179.2 (4) |
| N1—C6—C1—C2 | 177.8 (5) | C1—C2—C3—C4 | −0.9 (9) |
| N3—C13—C12—C11 | 177.5 (5) | C11—C10—C9—C8 | 0.8 (9) |
| N3—C13—C8—O5 | −0.2 (5) | C14—O5—C8—C13 | −0.2 (5) |
| N3—C13—C8—C9 | −178.1 (5) | C14—O5—C8—C9 | 177.5 (5) |
| C5—C6—C1—O4 | −179.9 (4) | C14—N3—C13—C12 | −177.7 (5) |
| C5—C6—C1—C2 | −1.0 (8) | C14—N3—C13—C8 | 0.6 (5) |
| C5—C4—C3—C2 | 0.5 (10) | C8—O5—C14—N3 | 0.6 (5) |
| C6—N1—C7—O4 | −0.1 (5) | C8—O5—C14—N4 | 178.4 (4) |
| C6—N1—C7—N2 | −179.8 (5) | C8—C13—C12—C11 | −0.7 (7) |
| C6—C5—C4—C3 | −0.3 (8) | C4—C5—C6—N1 | −177.9 (5) |
| C6—C1—C2—C3 | 1.1 (8) | C4—C5—C6—C1 | 0.5 (7) |
Symmetry code: (i) −x+1, y, −z+3/2.
Tetrakis(2-aminobenzoxazole-κN1)bis(nitrato-κO)cadmium(II) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N4—H4A···O1i | 0.86 | 2.26 | 2.971 (5) | 140 |
| N4—H4B···O2ii | 0.86 | 2.28 | 2.822 (6) | 121 |
| N2—H2A···O2i | 0.86 | 2.11 | 2.899 (7) | 152 |
| N2—H2B···O3iii | 0.86 | 2.33 | 2.953 (6) | 129 |
Symmetry codes: (i) −x+1, y, −z+3/2; (ii) x+1/2, y−1/2, z; (iii) x+1/2, y+1/2, z.
References
- Ali, S., Omprakash, P., Tengli, A. K., Mathew, B., Basavaraj, M. V., Parkali, P., Chandan, R. S. & Kumar, A. S. (2022). Polycyclic Aromat. Compd.30, 3853–3886.
- Clayden, J., Greeves, N., Warren, S. & Wothers, P. (2001). In Organic Chemistry. Oxford University Press.
- Decken, A. & Gossage, R. A. (2005). J. Inorg. Biochem.99, 664–667. [DOI] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst.42, 339–341.
- Erol, M., Celik, I., Uzunhisarcikli, E. & Kuyucuklu, G. (2022). Polycyclic Aromat. Compd.42, 1679–1696.
- Fan, L., Luo, Z., Yang, C., Guo, B., Miao, J., Chen, Y., Tang, L. & Li, Y. (2022). Mol. Divers.26, 981–992. [DOI] [PMC free article] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Imaizumi, T., Otsubo, S., Komai, M., Takada, H., Maemoto, M., Kobayashi, A. & Otsubo, N. (2020). Bioorg. Med. Chem.28, 115622. [DOI] [PubMed]
- Katritzky, A. R. & Pozharskii, A. F. (2000). In Handbook of Heterocyclic Chemistry, 2nd ed. New York: Academic Press.
- Khajondetchairit, P., Phuangsawai, O., Suphakun, P., Rattanabunyong, S., Choowongkomon, K. & Gleeson, M. P. (2017). Chem. Biol. Drug Des.90, 987–994. [DOI] [PubMed]
- Lynch, D. E. (2004). Acta Cryst. E60, o1715–o1716.
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst.53, 226–235. [DOI] [PMC free article] [PubMed]
- Ouyang, L., Huang, Y., Zhao, Y., He, G., Xie, Y., Liu, J., He, J., Liu, B. & Wei, Y. (2012). Bioorg. Med. Chem. Lett.22, 3044–3049. [DOI] [PubMed]
- Paramashivappa, R., Phani Kumar, P., Subba Rao, P. V. & Srinivasa Rao, A. (2003). Bioorg. Med. Chem. Lett.13, 657–660. [DOI] [PubMed]
- Parlapalli, A. & Manda, S. (2017). J. Chem. Pharm. Res.9, 57–62.
- Potashman, M. H., Bready, J., Coxon, A., DeMelfi, T. M., DiPietro, L., Doerr, N., Elbaum, D., Estrada, J., Gallant, P., Germain, J., Gu, Y., Harmange, J. C., Kaufman, S. A., Kendall, R., Kim, J. L., Kumar, G. N., Long, A. M., Neervannan, S., Patel, V. F., Polverino, A., Rose, P., van der Plas, S., Whittington, D., Zanon, R. & Zhao, H. (2007). J. Med. Chem.50, 4351–4373. [DOI] [PubMed]
- Razzoqova, S., Ibragimov, A., Torambetov, B., Kadirova, S., Holczbauer, T., Ashurov, J. & Ibragimov, B. (2023). Acta Cryst. E79, 862–866. [DOI] [PMC free article] [PubMed]
- Razzoqova, S., Torambetov, B., Amanova, M., Kadirova, S., Ibragimov, A. & Ashurov, J. (2022). Acta Cryst. E78, 1277–1283.
- Razzoqova, S., Torambetov, B., Todjiev, J., Kadirova, S., Ibragimov, A., Ruzmetov, A. & Ashurov, J. (2024). IUCrData9, x240033. [DOI] [PMC free article] [PubMed]
- Rigaku OD (2020). CrysAlis PRO. Rigaku Oxford Diffraction Ltd, Yarnton, England.
- Sangi, D. P., Meira, Y. G., Moreira, N. M., Lopes, T. A., Leite, M. P., Pereira–Flores, M. E. & Alvarenga, E. S. (2019). Pest Manage. Sci.75, 262–269. [DOI] [PubMed]
- Sattar, R., Mukhtar, R., Atif, M., Hasnain, M. & Irfan, A. (2020). J. Heterocycl. Chem.57, 2079–2107.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Šlachtová, V. & Brulíková, L. (2018). ChemistrySelect3, 4653–4662.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm11, 19–32.
- Spackman, M. A. & McKinnon, J. J. (2002). CrystEngComm4, 378–392.
- Spackman, P. R., Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Jayatilaka, D. & Spackman, M. A. (2021). J. Appl. Cryst.54, 1006–1011. [DOI] [PMC free article] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst.43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989025004049/wm5756sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989025004049/wm5756Isup2.hkl
CCDC reference: 2448896
Additional supporting information: crystallographic information; 3D view; checkCIF report
Additional supporting information: crystallographic information; 3D view; checkCIF report





