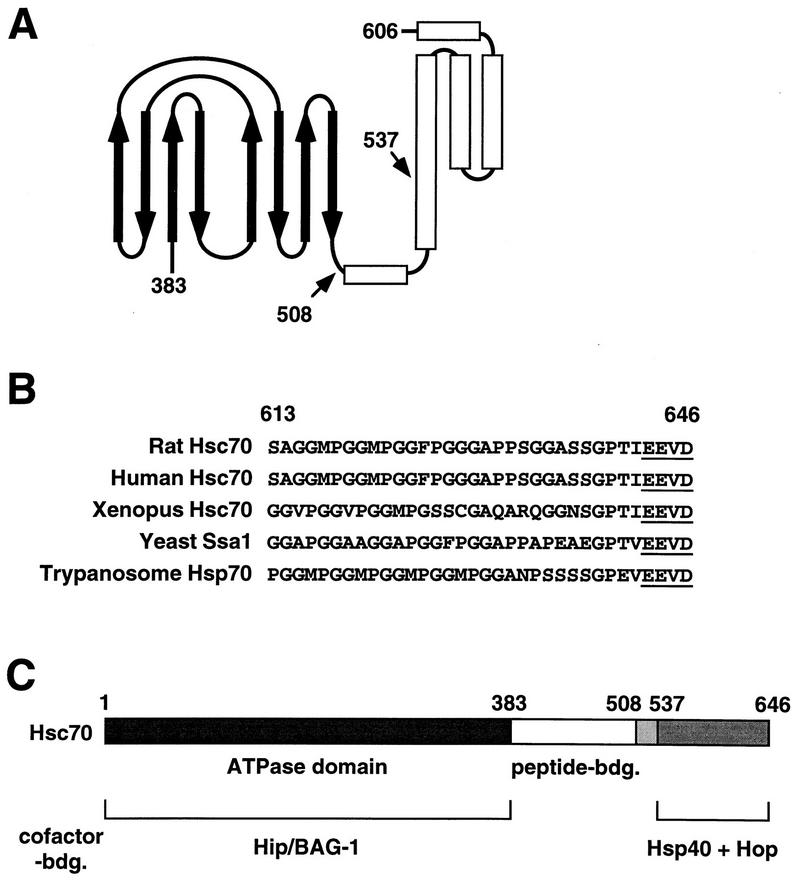
FIG. 8.
(A) Schematic representation of the peptide binding region of bacterial Hsp70. The peptide binding site is formed by two sheets of four antiparallel β-strands (thick arrows) connected by extended loops. The β sandwich is followed by five α helices (open bars) that appear to form a lid over the peptide binding site. Together, the carboxy-terminal half of the second helix and helices 3 to 5 form a tightly folded subdomain. The carboxy-terminal deletion fragments of Hsc70 used in this study start at positions 508 and 537 at the junction between the peptide binding site and the carboxy terminus (arrows). The numbers indicate the corresponding residues of rat Hsc70. The schematic representation does not include the GGMP repeats and the extreme carboxy terminus of Hsc70. (B) Multiple degenerate repeats of the tetrapeptide GGMP and the EEVD motif (underlined) are found at the carboxy terminus of eukaryotic cytosolic members of the Hsp70 protein family. The numbers indicate residues of rat Hsc70. (C) Domains of Hsc70 mediating interactions with chaperone cofactors. While Hip and BAG-1 bind to the ATPase domain of Hsc70 (amino acids 1 to 383) in a mutually exclusive manner, Hsp40 and Hop recognize distinct binding sites within the carboxy terminus of the chaperone protein (amino acids 537 to 646). bdg., binding.