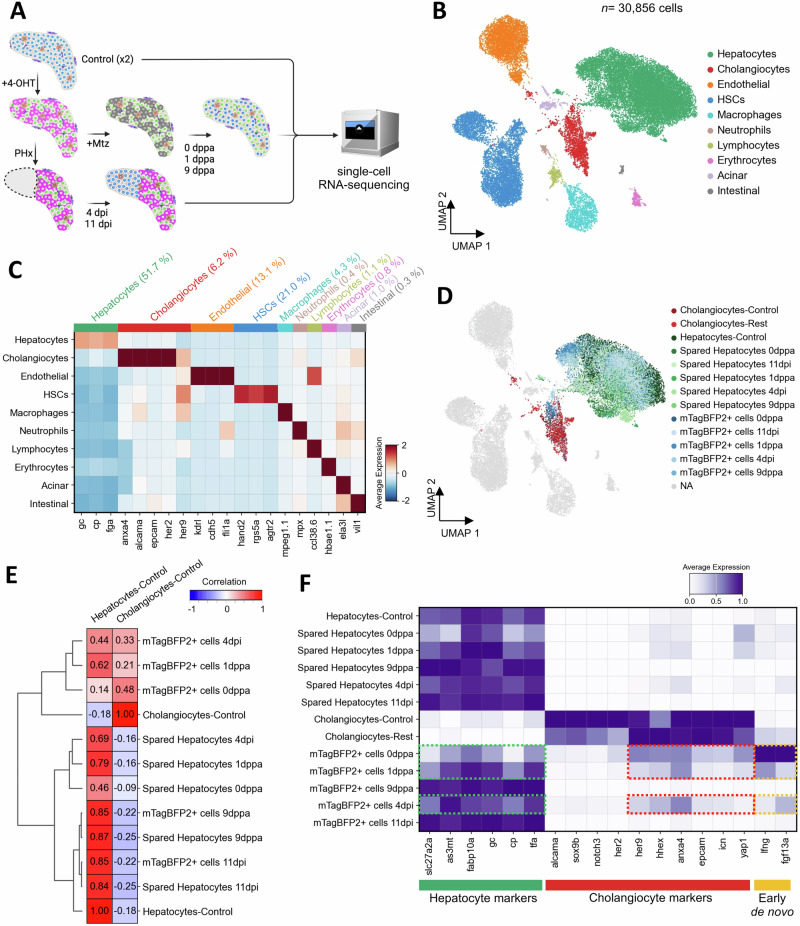
Fig. 5. De novo hepatocytes display transcriptional convergence with uninjured hepatocytes.
A Schematic representation of the single-cell RNA-seq approach to investigate the cellular and transcriptional landscape in basal condition (control-uninjured, N = 2) and during regeneration after partial ablation (0,1,9 dppa) and partial hepatectomy (4, 11 dpi). B UMAP visualization of cell types detected in zebrafish larval liver, combining data from injured and uninjured liver conditions. C Heatmap displaying the average scaled expression of selected markers across different cell types. D UMAP visualization focusing on specific injured conditions, highlighting mTagBFP2+ cells and spared hepatocytes (0,1,9 dppa and 4,11 dpi), control hepatocytes, control cholangiocytes, and cholangiocytes in injured conditions (cholangiocytes-rest). The remaining clusters are shown in gray. E Correlation matrix of hierarchically clustered mTagBFP2+ and spared hepatocytes compared to control hepatocytes and cholangiocytes. F Heatmap showing the normalized expression of hepatocyte and cholangiocyte-specific marker genes. Expression of selected genes that are detected in mTagBFP2+ cells at 0 dppa, 1 dppa and 4 dpi are outlined (dashed rectangles).