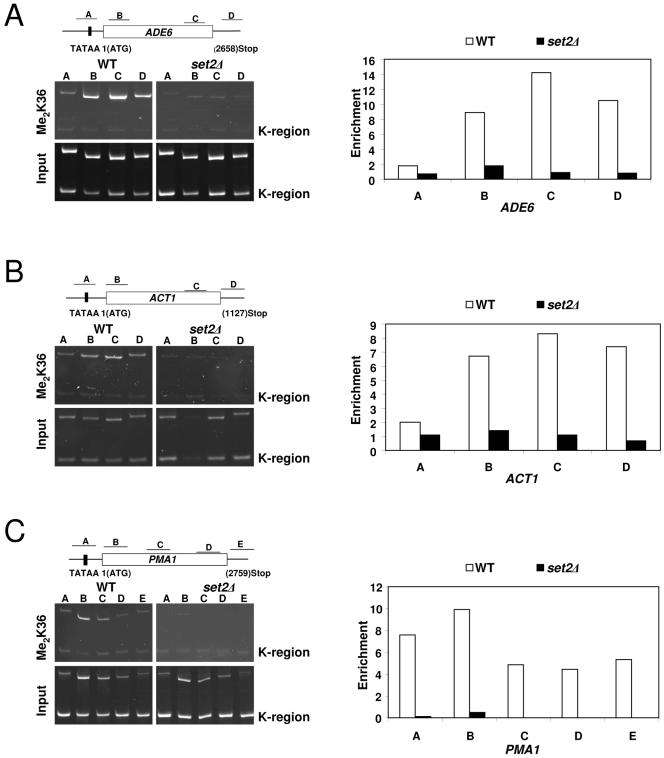
FIG. 5.
Set2-mediated K36 methylation is preferentially associated with the transcribed regions of active S. pombe genes. Left panels A to C: Chromatin immunoprecipitation assays were used to monitor the location of K36 dimethylation on actively transcribed genes (ADE6, ACT1, and PMA1) in wild-type and set2Δ strains using an H3 Me2K36-specific antibody. DNA from enriched precipitates (IP) were isolated and used in PCRs with promoter- and coding region-specific primer pairs for the indicated genes. A DNA fragment from the silent mating type loci (K region) of S. pombe known to lack modifications associated with active genes (H3 K4 methylation and H3 K14 acetylation) (34) was used as a control to normalize and calculate the relative enrichment of gene sequences in immunoprecipitated samples. Right panels: Quantification of the ChIP results shown in A to C. Relative enrichment values shown on the y axes were calculated by dividing the ratio of band intensities for immunoprecipitated DNA K region with the ratio of intensities for the input DNA K region. Gels and graphs are representative experiments from three independent repeats.