Abstract
Levonorgestrel is currently the most widely used contraceptive, with advantages that include rapid oral absorption and high bioavailability. Among the various synthetic methods employed in its production, Saccharomyces cerevisiae AS2.346 was first introduced by the Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, in 1976, successfully catalyzing the conversion of ethyl secodione to (13R,17S)-ethyl secol. Presently, Saccharomyces cerevisiae AS2.346 whole-cell catalysis is still used in levonorgestrel industrial production. To identify the enzyme responsible for catalyzing the conversion of ethyl secodione to (13R,17S)-ethyl secol, we isolated two genes, Assdr1 and Assdr2, both encoding carbonyl reductases. The biochemical characterization of AsSDR1 and AsSDR2 showed that they had high activity for ethyl secodione. Functional analysis demonstrated that the deletion of Assdr1 almost completely abolished the production of (13R,17S)-ethyl secol, while the deletion of Assdr2 had little to no effect on the production, indicating that Assdr1 plays a pivotal role in the biosynthesis of (13R,17S)-ethyl secol. The reason for the difference in the function of AsSDR1 and AsSDR2 in vivo may be attributed to their different cellular localizations, with AsSDR1 located outside the mitochondria and AsSDR2 in the mitochondria. A metabolomics analysis of Assdr1 knockout strains revealed that AsSDR1 may act as a pterin reductase. Overexpression of the newly isolated gene Assdr1 led to an average 47.88 % higher (13R,17S)-ethyl secol yield compared to that of the wild-type strain. Furthermore, engineering the metabolism of the NADPH cofactor by overexpressing the truncated gene pos5Δ17 encoding mitochondrial NADH kinase produced a 111.25 % higher (13R,17S)-ethyl secol yield compared to that of the wild-type strain. These findings not only elucidate the key enzymatic players involved in the synthesis of (13R,17S)-ethyl secol but also provide a framework for optimizing the industrial biotransformation process for levonorgestrel production.
Keywords: Carbonyl reductase, Saccharomyces cerevisiae AS2.346, Ethyl secodione, Metabolic engineering
1. Introduction
Steroidal drugs are the second-largest class of marketed drugs, after antibiotics. C17-hydroxylated steroids belong to a family of natural steroids with important medicinal value and are useful precursors for the synthesis of other high-performance steroid drugs [1]. Pharmacological studies have shown that the efficacy of steroidal hormone drugs is closely related to the C17 side chain. In the chemical synthesis of steroids, substitution reactions are usually used to protect the C17 side chain, resulting in an excessively lengthy synthetic process. At the same time, these reactions often require the use of strong acids or strong bases for catalysis, contributing to environmental concerns.
Currently, biotransformation is widely used in industrial production due to its environmental friendliness. A valuable biocatalyst should have the characteristics of a high catalytic conversion rate and high selectivity, including enantioselectivity, regioselectivity, and chemoselectivity, as well as high process stability under the conditions required for chemical transformation. Kristan et al. [2] performed site-directed mutagenesis of C17 carbonyl reductase in the filamentous fungus Cochliobolus lunatus, which altered substrate specificity and coenzyme requirements, and improved overall catalytic activity. Point mutations in the substrate-binding region Val161Gly and Tyr212Ala increased the initial rate of conversion of androstenedione to testosterone. Zhou et al. [3] improved the catalytic activity of the stereotactic reduction of androstenedione 17β by adjusting the flexibility of the α6-helix at the entrance of the RasADH substrate without sacrificing enantioselectivity. Compared with the wild-type enzyme, the catalytic efficiency of the mutants F205I and F205A increased by 623-fold and 523-fold, respectively.
The biotransformation reaction in this study belongs to the C17 hydroxylation reaction of steroids. As the most widely used oral contraceptive, levonorgestrel is the levorotatory form of norgestrel, a synthetic progestogen with progestational and androgenic activity [4], which suppresses luteinizing hormone activity, inhibits ovulation [5], and induces changes in the cervical mucus and endometrium [6]. Due to the problems with its chemical synthesis [7] (Fig. S1) such as a long synthesis route, low productivity, and serious environmental pollution, along with the formation of racemic products, scientists are turning to biosynthetic methods to produce chiral intermediates. These intermediates are then further processed via chemical synthesis to yield levonorgestrel [8]. (Fig. S2). The Saccharomyces cerevisiae AS2.346 strain [9], successfully catalyzes the intermediate ethyl secodione into (13R,17S)-ethyl secol (Fig. 1a). Presently, whole-cell catalysis is still used in industrial production. However, the genome of Saccharomyces cerevisiae AS2.346 has not been reported. It is still unknown which protein performs the transformation.
Fig. 1.
Identification, characterization of AsSDR1 and AsSDR2. (a) Saccharomyces cerevisiae AS2.346 transformation reaction; (b) Phylogenetic analysis of carbonyl reductases. The phylogenetic tree was constructed based on the alignment of full amino acid sequences. All analyzed sequences of reductase enzymes were retrieved from GenBank and SGD; (c) Biotransformation reaction of recombination E. coli.
Recently, some carbonyl reductase were also found to be an efficient (13R,17S)-ethyl secol producer. Contente et al. [10] identified an enzyme with asymmetric reducing ability in Pichia glucozyma CBS 5766, KRED1-Pglu. Subsequently, they cloned the gene into E. coli BL21(DE3) to test its reducing ability. In a 1-L reaction system, 2.5 % KRED1-Pglu converted 6.5 mmol/L substrate within 4 h with a 65 % yield. Chen et al. [11] generated 14,17-dihydroxyl reduction products when using the asymmetric reductase RasADH (wild type) from Ralstonia sp. After site-directed mutagenesis of RasADH, a mutant strain F12 (I91V/I187S/I188L/Q191N/F205A) with a monohydroxyl reduction yield of 99.5 % at position 17 and a 14,17-dihydroxyl reduction yield of 0.1 % was obtained, which showed a 183-fold enhancement of enzyme activity. In 2020, TbADH, a carbonyl reductase from Thermoanaerobium brockii, was used to transform ethyl secodione [12]. The pET21a(+)-N114L/M285L/L294P-BL21 strain with improved viability and optimal stereoselectivity was successfully screened with a point mutation in TbADH. In 100 mL of sodium phosphate buffer (pH 7.5, 100 mM), 30 g/L of wet cells and 20 mL of isopropanol containing the substrate were added to reach a final substrate concentration of 150 g/L, with NADP+ at a final concentration of 0.2 g/L. The reaction was carried out at 37 °C and 200 rpm. After 36 h, the substrate conversion rate reached 99 %.
In order to obtain more insights into the carbonyl reductase in AS2.346 and with an aim to further improve (13R,17S)-ethyl secol production, we identified and characterized an additional carbonyl reductase (AsSDR1) in AS2.346. Overexpression of this gene together with the truncated pos5Δ17, which allowed NADPH to be rapidly replenished, yielded significantly increased (13R,17S)-ethyl secol productivity.
2. Materials and Methods
2.1. Chemicals
Ethyl secodione and (13R,17S)-ethyl secol were obtained from China Resources Zizhu Pharmaceutical Co. (Beijing, China). Geneticin and kanamycin sulfate was obtained from BBI Life Sciences (Shanghai, China), while yeast extract and tryptone were obtained from OXOID (Beijing, China). All other chemicals were obtained from Yongda Chemical (Tianjin, China) and were of analytical grade.
2.2. Strains, media, and culture conditions
The Escherichia coli and Saccharomyces cerevisiae strains used in this study are listed in Table S1. E. coli was cultured at 37 °C in LB medium with 25 mg/L kanamycin for selection. S. cerevisiae was grown at 28 °C in YPD (10 g/L yeast extract, 20 g/L tryptone, 20 g/L glucose), with 200 μg/mL geneticin added when needed for transformant selection.
The shake flask culture method was used, as described below. A single colony was inoculated into 20 mL YPD and grown at 30 °C, 220 rpm. The cells were transferred to 50 mL of YPD medium at a 5 % ratio and cultured for 24 h at 28 °C and 220 rpm. Then, 3 mL of a 5 g/L substrate ethanol solution was added to the 50-mL reaction system. The cells were incubated for 96 h at 28 °C and 220 rpm. At the end of the reaction, the mixture was extracted with an equal volume of ethyl acetate, shaken at 28 °C and 220 rpm for 10 min, and centrifuged at 10,000 rpm at room temperature for 10 min. The supernatant was used for HPLC analysis.
2.3. Analytical methods and (13R,17S)-ethyl secol production
Ethyl secodione conversion and (13R,17S)-ethyl secol production were determined by HPLC analysis using a Welch Ultimate AQ-C18 column (4.6 × 250 mm). A mixture of methanol/ddH2O (v/v, 70/30) was used as the eluent solvent at a flow rate of 1 mL/min, with ultraviolet detection at 240 nm and column temperature maintain at 34 °C. The retention times were 13.6 min for ethyl secodione and 10.8 min for (13R,17S)-ethyl secol.
2.4. General molecular biological techniques
Standard media and techniques used for E. coli were used as described in Molecular Cloning [13]. The media and techniques used for Saccharomyces cerevisiae were previously described in Methods in Yeast Genetics [14]. The restriction enzymes, DNA polymerases, and ligase were supplied by Thermo Fisher Scientific (Waltham, MA, USA). Genomic DNA from Saccharomyces cerevisiae was prepared using a method consistent with the previously described procedures [13]. PCR was performed using the primers listed in Table S1. Pfu DNA polymerase (Sangon Biotech, Shanghai, China) was used for cloning, while rTaq DNA polymerase (Takara, Beijing, China) was used to verify the gene integration into the genome. The PCR fragments were purified from agarose gels using a GeneJet gel extraction kit (Thermo Scientific). Primer synthesis was performed by Genscript (www.genscript.com.cn). DNA sequencing was performed by Genewiz (www.genewiz.com.cn). Yeast transformation was performed as described in Cold Spring Harbor protocols [15].
2.5. Sequence analysis
The experimental methods employed have been described in previous studies [16]. All analyzed sequences of carbonyl reductase enzymes were retrieved from GenBank and the Saccharomyces Genome Database (www.yeastgenome.org).
2.6. Cloning of carbonyl reductase-encoding genes and mutants
The KRED1-Pglu sequence (GenBank: KR080472.1) was synthesized by Changzhou Jiyu Biological. PCR amplification from strain HCY000 genomic DNA used primers (Table S1) with NdeI/XhoI sites. Amplicons were digested and cloned into pET28a, then transformed into E. coli BL21(DE3). Mutations were generated by overlap extension PCR, and constructs were verified by sequencing (T7 primer, Genewiz).
2.7. Construction of AsSDR1 and AsSDR2 disruption strain
Assdr1 or Assdr2 were deleted in Saccharomyces cerevisiae AS2.346 using CRISPR–Cas9 technology. The guide RNA plasmid pCAS-AsSDR1/pCAS-AsSDR2 (Table S1) was amplified from pCAS as a template using the primer pair pCAS-U and pCAS-D (Table S1) carrying a 20-bp PAM sequence (pCAS-AsSDR1-2/pCAS-AsSDR1-3//pCAS-AsSDR2-2/pCAS-AsSDR2-3) for the Assdr1 or Assdr2 deletion. The plasmids were used to transform Saccharomyces cerevisiae AS2.346. Transformants were selected using geneticin (200 μg/mL) as a selectable marker. The sequence of the gene disruption was verified by analytical PCR using the primer pairs AsSDR1-U/AsSDR1-D and AsSDR2-U/AsSDR2-D (Table S1). The sequences were also verified by DNA sequencing in genewiz to determine whether a stop codon mutation had been created. The resulting strains were designated HCY100 (disruption of AsSDR1), HCY200 (disruption of AsSDR2), and HCY300 (disruption of AsSDR1 and AsSDR2).
2.8. Complementation of disruption strains
The promoter TDH3 and terminator CYC1 were amplified with primer pairs pTDH3-U1/pTDH3-D1 and tCYC1-U1/tCYC1-D1 (Table S1), respectively. Genes Assdr1 and Assdr2 were amplified with primer pairs AsSDR1-U2/AsSDR1-D2 and AsSDR2-U2/AsSDR2-D2 (Table S1), respectively. These fragments were ligated (using C115 ligase) into BamHI/XmaI-digested pRS303K-18s (a pRS303K derivative for 18S rDNA integration, modified with primers 18s-1 to 18s-4). The resulting constructs were designated pRS303K-18s-AsSDR1 and pRS303K-18s-AsSDR2, respectively (Table S1). The constructed plasmid was digested with NaeI/AatII and then transformed into the HCY300 strain. Transformants were selected on YPD medium supplemented with geneticin (200 μg/mL) and verified by PCR using primers Y–18S-1 to Y–18S-4. The resulting strains were designated HCY301 (complementation of AsSDR1) and HCY302 (complementation of AsSDR2).
2.9. Subcellular localization of AsSDR1 and AsSDR2
Plasmid pRS303K-18s-EGFP, pRS303K-18s-AsSDR1-EGFP and pRS303K-18s-AsSDR2-EGFP were constructed in the same manner as the complementation plasmid except that different primers were used. The resulting strains were designated HCY400 (expressing EGFP), HCY401 (expressing AsSDR1-EGFP), and HCY402 (expressing AsSDR2-EGFP).
An appropriate amount of yeast cells was washed twice with 1 mL PBS. The cells were centrifuged at 12,000 rpm for 20 s, and the supernatant was removed. Then, 1 mL of 70 % ethanol was added to fix the cells for 20 min. After centrifugation at 12,000 rpm for 20 s, the supernatant was removed, washed twice with 1 mL PBS, and resuspended in 100 μL of PBS. A 10 μL of cell suspension droplet was placed on a glass slide, followed by the addition of 5 μL of antifluorescence quencher. The fluorescence localization was observed through an oil immersion lens (magnification 100×) and photographed.
2.10. HCY100 metabolomics analysis
Following the protocol in Materials and Methods 2.2, yeast cells were harvested post-fermentation and analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS). After preprocessing the data, the metabolomics data were further analyzed using principal component analysis (PCA), partial least squares discriminant analysis (PLS-DA) and orthogonal partial least squares-discriminant analysis (OPLS-DA). Differential metabolites between the HCY000 and HCY100 groups were identified, and the pathways associated with these differential metabolites and their networks were analyzed. Potential metabolite markers that were significantly altered were further identified.
2.11. Protein production and crude enzyme reaction
Escherichia coli BL21(DE3) cells expressing wild-type and mutant genes were cultivated overnight at 37 °C. A 1 L culture was induced with 0.1 mM IPTG at OD600 = 0.6 and incubated for 20 h at 16 °C. The samples were centrifuged at 10,000 rpm for 10 min, and the supernatant was collected for HPLC analysis. Cells were centrifuged, resuspended in phosphate buffer (100 mM, pH 8.0), and sonicated. After centrifugation (10,000 rpm, 20 min), the filtered extract was reacted with 200 μL ethanol substrate (50 g/L) at 37 °C for 3 h. The solution was extracted with ethyl acetate, centrifuged, and analyzed by HPLC.
2.12. Protein purification and enzyme reaction
Cells were lysed as above, and AsSDR1/AsSDR2 were purified using Ni-NTA agarose (elution buffer: 20 mM Tris, 500 mM NaCl, 250 mM imidazole, pH 8.0). SDS-PAGE (8 % gel) and Coomassie staining were performed, with protein concentration determined via BSA standard curve.
The activities of AsSDR1 and AsSDR2 were determined by HPLC. The effect of temperature on AsSDR1 and AsSDR2 was analyzed at 20–50 °C (in 100 mM Tris-HCl buffer, pH 8.0), while the pH effect was tested at 3.0–9.0 (using different buffer systems) at 30 °C. The reaction mixture contained 10 U GDH, 25 mM glucose, 0.1 mM NADPH, 0.5 mg substrate, and 100 μL carbonyl reductase (1 mg/mL). All assays were performed in triplicate.
2.13. Molecular docking simulation
The enzyme protein model was obtained from the PDB library (PDB ID:6UHX) (https://www.rcsb.org/). The substrate model was constructed by Chemdraw19 and optimized by Desmond V6.6 for energy minimization. The molecular docking was performed by AutoDock Vina V1.1.2, using the docking conformation with the lowest energy. Desmond V6.6 was used to conduct the molecular dynamics simulation. For the simulation, the protein was placed in a 1-nm cube using the AMBER14SB force field. The water in the box solvent model used the TIP3P water model as the solvent, and by adding Na+ and Cl− at 0.15 mmol/L, the charge balance of the entire box of the solution system was achieved. The pH was set at 7.4. The conjugate gradient method was applied to the entire system first to minimize the energy using 1000 steps. Under the canonical ensemble, the system was heated from 0 to 303.15K, with an integration step of 1 fs, over a duration of 100 ps. The system was then brought to equilibrium for 50 ps, and the protein ligand complex was simulated for 100 ns. The protein structure results were analyzed by Pymol V2.4, and the graph of the correlation distance was analyzed by Origin 2019.
2.14. Overexpression of carbonyl reductases encoding the gene mutant AsSDR1 I180A
The YARCdelta5 sites were selected as intergenic regions for integrating expression cassettes using Cas9-based genome editing. The YARCdelta5 site is a long terminal repeat sequence located in chromosome I. Based on the targeting sites, guide RNA-expressing plasmids pCAS-δ5 were constructed by overlap PCR using primers pCAS-U/pCAS-D and pCAS-δ5-2/pCAS-δ5-3. The gene cassette pTDH3-AsSDR1 was amplified using the primer pair δ5-1 and δ5-4 as donor DNA for CRISPR–Cas9-based integration (Table S1). Plasmids pCAS-δ5 and gene cassette pTDH3-AsSDR1 were then used to transform HCY000. Transformants were selected on YPD medium supplemented with geneticin (200 μg/mL) and were confirmed by PCR using primers Y-δ5-1 and Y-δ5-4. The resulting strain was designated HCY501.
2.15. Overexpression of the truncated gene pos5Δ17
The CS6 sites [17] were selected as intergenic regions for integrating expression cassettes via Cas9-based genome editing. The CS6 site is a long terminal repeat sequence located in chromosome VII between YGR190C and tW(CCA)G2. Based on the targeting sites, guide RNA-expressing plasmids pCAS-CS6 were constructed by overlap PCR using primers pCAS-U/pCAS-D and pCAS-CS6-2/pCAS-CS6-3. The gene cassette pTEF1-POS5Δ17 was amplified using the primer pair CS6-1 and CS6-4 as donor DNA for CRISPR–Cas9-based integration (Table S1). Plasmids pCAS-CS6 and gene cassette pTEF1-POS5Δ17 were then used to transform HCY501. Transformants were selected on YPD medium supplemented with geneticin (200 μg/mL) and were confirmed by PCR using primers Y-CS6-1 and Y-CS6-4. The resulting strains were designated HCY502.
2.16. RNA isolation and transcript quantification
Cells were collected at an OD600 of 2.0. Total RNA was extracted using a SparkZol kit from SparkJade (Jinan, China), and cDNA was obtained using a PrimeScript™ RT reagent kit with gDNA eraser (Takara, Dalian, China). qRT-PCR was performed using SYBR Green qPCR Mix (Biosharp, Hefei, China) on a QuantStudio3 (Thermo Fisher Scientific, Waltham, MA, USA). Primer pairs for qRT-PCR were rt-AsSDR1-U/rt-AsSDR1-D, rt-AsSDR2-U/rt-AsSDR2-D, and rt-POS5-U/rt-POS5-D for genes Assdr1, Assdr2 and pos5Δ17, respectively (Table S1). Gene expression was normalized to that of the actin gene (yfl039c, primers rt-β-actin-U/rt-β-actin-down) using the ΔCT method. The fold differences in gene expression between the transformants and the control strain HCY000 were calculated as 2−ΔΔCT [18]. All samples were analyzed in triplicate.
3. Results and discussion
3.1. Identification of carbonyl reductase-encoding genes in Saccharomyces cerevisiae AS2.346
It has been reported [10] that the recombinant ketoreductase KRED1-Pglu was used in the ethyl secodione biotransfomation reaction at a 1–2 g/L substrate concentration, achieving a 65 % conversion rate. This carbonyl reductase, belonging to the short-chain dehydrogenase/reductase family (SDR), has also been reported to be dependent on NADPH as a redox co-factor.
Saccharomyces cerevisiae AS2.346 (HCY000) was used by the Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences in 1976 to successfully catalyze ethyl secodione into (13R,17S)-ethyl secol. Currently, whole-cell catalysis is still used in industrial production. However, the existence of multiple carbonyl reductases complicates the biotransformation. The genome of HCY000 has not been reported, while the genome of the standard strain S. cerevisiae S288C (NC_001141.2) has been reported. Therefore, the S. cerevisiae S288C genome was used for BLAST analysis in the next step.
A protein BLAST search using KRED1-Pglu as a query highlighted the Saccharomyces cerevisiae S288C genes yir035c and yir036c with a 45.2 and 47.49 % identity, respectively. A BLAST analysis of the RasADH protein sequence with YIR035C and YIR036C showed a 27.92 and 28.65 % identity, respectively. YIR035C and YIR036C belong to the SDR superfamily, and their protein sequences share 51.33 % identity. This analysis showed that YIR035C and YIR036C were completely consistent with KRED1-Pglu and RasADH in terms of the position of their active residues, and it was speculated that their functions were similar to those of KRED1-Pglu and RasADH, with an ethyl secodione reducing ability. To verify the BLAST analysis, a phylogenetic tree (Fig. 1b) was also constructed with a full-length amino acid sequence of KRED1-Pglu, RasADH, YIR035C and YIR036C using MEGA7 software. The phylogenetic tree indicates that YIR035C and YIR036C originating from Saccharomyces cerevisiae S288C are more closely related to KRED1-Pglu from Ogataea, while being more distantly related to RasADH from Ralstonia. Primers were designed based on the sequences of YIR035C and YIR036C from Saccharomyces cerevisiae S288C to identify their homologous sequences in Saccharomyces cerevisiae AS2.346. Since yir035C and yir036C follow systematic gene nomenclature—representing the 35th and 36th open reading frames (ORFs) on the right arm of chromosome I in the yeast genome—and the genome of AS2.346 remains uncharacterized. A standard naming convention was adopted for the identified sequences, combining the strain abbreviation, gene function and numerical identifiers: Assdr1 and Assdr2. Therefore, it was hypothesized that the two genes were capable of undergoing transformation reactions. In order to confirm the predicted catalytic activity, Assdr1 and Assdr2 were PCR amplified from the genomic DNA of AS2.346 using primers listed in Table S1. The amplified fragments were cloned into a pET28a vector and expressed in E. coli BL21(DE3). Assays evaluating the reductase activity of IPTG-induced cells using ethyl secodione as a substrate led to the identification of HCE010 (gene Assdr1, AsSDR1) and HCE020 (gene Assdr2, AsSDR2) with NADPH-dependent carbonyl reductase activity. The reduction of ethyl secodione into (13R,17S)-ethyl secol was detected in the cells of strains HCE010 and HCE020, as shown in Fig. 1c and Fig. S3. No reductase activity was found in the non-transformed E. coli or HCE000 strain (gene kred, KRED1-Pglu) under the experimental conditions.
3.2. Disruption of two carbonyl reductase-encoding genes and their effects on (13R,17S)-ethyl secol synthesis
To confirm that the two putative identified carbonyl reductases were involved in (13R,17S)-ethyl secol biosynthesis, the corresponding genes (Assdr1 and Assdr2) were disrupted in HCY000. The disruption plasmids, pCAS-AsSDR1 and pCAS-AsSDR2, were constructed as described in the Methods section and used to disrupt the corresponding genes separately. The disrupted mutants were grown in YPD medium and ethyl secodione was added, as described in the Methods section. As shown in Table 1, the specific growth rate of the disrupted mutant HCY100 significantly decreased from that of the parental strain. Compared to the parental strain HCY000, disruption of the gene Assdr1 (AsSDR1, strain HCY100) resulted in little transformation of ethyl secodione, whereas transformation with disruption of the gene Assdr2 (AsSDR2, strain HCY200) produced almost no change. From this, it was speculated that the gene encoding protein AsSDR1 was involved in steroid microbial conversion in HCY000.
Table 1.
(13R,17S)-Ethyl secol production by strain AS2.346 and E.coli.
| Strains | Yield of (13R,17S)-ethyl secol (%) | Yield of 14,17-dihydroxyl (%) |
|---|---|---|
| AS2.346 | 68.47 ± 3.15 | 16.40 ± 0.22 |
| HCY100 | 7.28 ± 0.22 | – |
| HCY200 | 59.53 ± 3.80 | 13.77 ± 2.27 |
| HCY300 | – | – |
| HCY301 | 41.99 ± 12.09 | 12.72 ± 3.28 |
| HCY302 | 27.56 ± 7.43 | 20.30 ± 2.40 |
| HCE010 | 71.40 ± 2.05 | 25.10 ± 5.17 |
| HCE011 | 82.54 ± 3.60 | 11.82 ± 7.99 |
| HCE012 | 98.26 ± 2.99 | – |
| HCE013 | 97.61 ± 5.14 | – |
| HCE014 | 28.83 ± 1.52 | – |
The values provided are the means of three independent replicates.
3.3. Complementation of the disruption strain
To further verify the roles played by AsSDR1 and AsSDR2 in HCY000, the double knockout strain HCY300 was constructed. Subsequently, two genes were back-patched to construct strains HCY301 (complementation AsSDR1) and HCY302 (complementation AsSDR2). A schematic diagram of the complementary strains is shown in Fig. S4. As shown in Table 1, compared to the parental strain HCY300, both HCY301 and HCY302 recovered production of (13R,17S)-ethyl secol. However, the yield of HCY301 was slightly higher than that of HCY302. Real-time fluorescence quantitative PCR (q-PCR) showed that the transcription level of AsSDR1 in HCY301 was approximately the same as that of AsSDR2 in HCY302 (Fig. S5). It was speculated that there may be other differences between these two proteins that produced the differences in yields.
3.4. Molecular dynamics simulations of AsSDR1 and AsSDR2
The function of a protein is closely related to its structure. The structure of AsSDR1 and AsSDR2 were analyzed to further determine the reasons for the difference in yield in vitro versus in vivo. The crystal structure of YIR035C (AsSDR1 homologous proteins) has been reported. However, there is no reported crystal structure of AsSDR2 or its homologous protein YIR036C. Therefore, AlphaFold (https://alphafold.com/) was used to construct a model of AsSDR2 in this experiment. The constructed AsSDR2 model was comprehensively evaluated online using the SAVES website (https://saves.mbi.ucla.edu/). The results showed that the AsSDR2 homologous model was sufficiently credible. The ERRAT score was 97.65 (Fig. S6a), and the residue distribution in the core area was 94.3 % (Fig. S6b) in the Ramachandran plot of the PROCHECK evaluation.
The RMSD and MD distances of AsSDR1 and AsSDR2 were analyzed (Fig. 2a and Fig. S7). The average attack distances of AsSDR1 and AsSDR2 were 3.56 Å and 5.02 Å, respectively. The protein-substrate binding pattern diagram (Fig. 2b and c) from the same perspective shows an attack distance between the H on the nicotinamide ring and the carbonyl carbon at substrate C17 in NADPH at a point in the simulation. The attack ranges of AsSDR1 and AsSDR2 were 3.6 Å and 3.8 Å, respectively. The effective distance of AsSDR2 was greater than that of AsSDR1, and was not conducive to a reaction occurring.
Fig. 2.
Molecular Dynamics Simulation Results of AsSDR1 and AsSDR2. (a) Protein-substrate MD distance of AsSDR1 and AsSDR2; (b) diagram of the protein-substrate binding patterns of AsSDR1; (c) diagram of the protein-substrate binding patterns of AsSDR2.
To further analyze the differences between AsSDR1 and AsSDR2, literature review and bioinformatics analysis were carried out. It was shown [19] that in S. cerevisiae 288C, the half-life of YIR035C (AsSDR1 homologous protein) was 11.9 h, and the half-life of YIR036C (AsSDR2 homologous proteins) was 10.8 h. The difference in the half-lives of the two proteins may greatly affect the amount of protein in S. cerevisiae, resulting in differences in yield.
When the transmembrane structures of the two proteins were predicted using PHOBIUS (https://phobius.sbc.su.se/), it was found that AsSDR1 was a free cytoplasmic protein, while AsSDR2 had a membrane-bound domain at leucine 111–130 (Fig. S8). This caused AsSDR2 to bind substrates less efficiently than AsSDR1, resulting in yield differences in the double-knockout single back-patched strains.
In addition, bioinformatics analysis of AsSDR1 and AsSDR2 proteins revealed that their locations in HCY000 may differ slightly.
3.5. Subcellular localization of AsSDR1 and AsSDR2
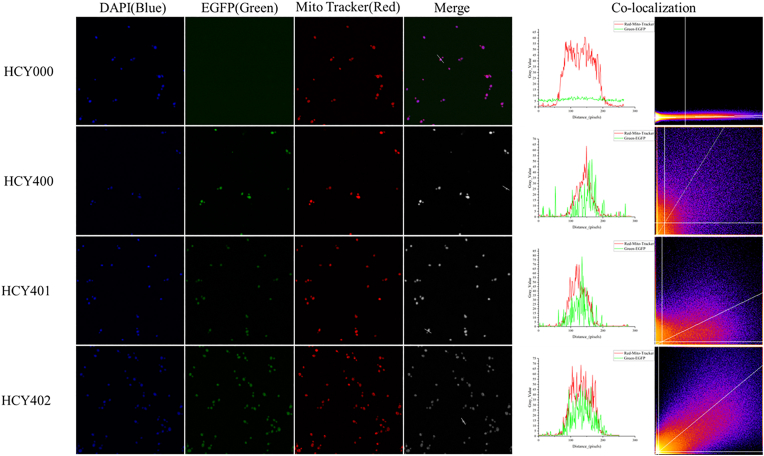
To confirm the differences in subcellular localization, AsSDR1 and AsSDR2 were fused to enhanced green fluorescent protein (EGFP) using fusion reporter gene mapping. The location of AsSDR1 and AsSDR2 in the cell was determined by fluorescence microscopy. Strain HCY400, HCY401, and HCY402 were constructed as described in the Methods section.
HCY000, HCY400, HCY401, and HCY402 were observed using fluorescence microscopy (Fig. 3). The intracellular DNA were labeled with the blue fluorescent dye DAPI (Beyotime Biotechnology). Mitochondria were labeled with the red fluorescent dye Mito-Tracker Red (Beyotime Biotechnology). The blue fluorescence (excitation wavelength: 350 nm, emission wavelength: 470 nm), red fluorescence (excitation wavelength: 587 nm, emission wavelength: 615 nm), green fluorescence (excitation wavelength: 488 nm, emission wavelength: 507 nm), and cell morphology under visible light were observed using an oil immersion lens (magnification × 100 times). Fluorescence pixel plots were generated using ImageJ software. Pearson's correlation coefficient (PCC) of the green and red channels was calculated using the Color 2 plugin (https://imagej.net/plugins/coloc-2).
Fig. 3.
Fluorescence microscopy was used to observe HCY000, HCY400, HCY401, and HCY042 strains. Blue is DAPI; green is the expressed EGFP protein; red is Mito-Tracker dye; merge is an overlay image; co-localization is a fluorescent pixel plot and 2D intensity histogram.
PCCs of HCY000, HCY400, HCY401, and HCY402 were 0.000, 0.3880, 0.2981 and 0.6050, respectively. PCCs of HCY000, HCY400, and HCY401 were less than 0.5, indicating that the correlation between the location of EGFP or AsSDR1-EGFP expression and mitochondria was weak, while the PCC of HCY402 was greater than 0.5, indicating that there was a certain degree of co-localization between AsSDR2-EGFP and mitochondria. Therefore, the differential subcellular localization of AsSDR1 (non-mitochondrial) and AsSDR2 (mitochondrial) provides a plausible explanation for their distinct behaviors: while exhibiting comparable expression yields in vitro, they demonstrate significantly different production levels when knocked out and complemented in Saccharomyces cerevisiae AS2.346.
3.6. Metabolomics analysis of HCY100
During the fermentation of strain HCY100, it was found that, compared with HCY000, HCY100 showed delayed growth and weakened fermentation flavor (Fig. S9). The reason for this phenomenon is unknown due to the lack of studies on the function of Assdr1 in HCY000. This study aimed to elucidate the underlying causes for this phenomenon through metabolomics analysis.
According to the base peak chromatogram (Fig. S10), the metabolites of the knockout strain HCY100 had changed to a certain extent compared to those of HCY000. PCA, PLS-DA and OPLS-DA were used to analyze the experimental data (Figs. S11 and S12, Table S2). There was a large difference between the two groups, and the samples within the groups were closely clustered. The results indicated that the reproducibility of the experiment was good, with a significant metabolic difference between the two groups (Fig. S13).
A total of 335 metabolites were identified, including 180 differential metabolites in total, with 70 upregulated differential metabolites and 110 downregulated differential metabolites (Table S3). The results suggested that these potential markers influenced metabolic pathways such as central carbon metabolism, ABC transporters, and protein digestion and absorption (Fig. S14, Table S4).
The metabolic disturbances observed in HCY100 included impaired carbon source utilization and a loss of key fermentation flavor compounds. Notably, primary metabolites such as 2,3-butanediol, γ-aminobutyric acid, malic acid, glutaric acid, sorbitol, hydroxypyruvate, and benzaldehyde showed significant changes. These alterations suggested that the disruption of Assdr1 affected the primary metabolism in HCY000, which, in turn, impacted the production of secondary metabolites like higher alcohols and esters, which are critical to the fermentation flavor profile. These findings point to the potential involvement of Assdr1 in the regulation of primary metabolic processes within HCY000.
Further analysis revealed substantial changes in the levels of various amino acids and their derivatives, including threonine, proline, arginine, phenylalanine, and tryptophan. Specifically, the content of tryptophan was reduced, while phenylalanine levels were elevated in the knockout strain HCY100, suggesting that Assdr1 plays a role in maintaining the balance of aromatic amino acid metabolism. The data imply that Assdr1 may be involved in the biosynthesis of aromatic amino acids or the metabolism of related cofactors.
Sequence alignment and structural analysis indicated that AsSDR1 shares high homology with human sepiapterin reductase (SPRE_HUMAN, P35270.1). The Pfam database search identified the SPR family (IPR006393), and comparison of AsSDR1 and AsSDR2 sequences with known functional sepiapterin reductases revealed that the active site residues of AsSDR1 and AsSDR2 are nearly identical to those of SPR, suggesting a similar enzymatic function. This led to the hypothesis that AsSDR1 and AsSDR2 may possess sepiapterin reductase activity, capable of reducing sepiapterin in a manner analogous to human SPR (Fig. S15).
These results provide valuable insights into the metabolic effects of Assdr1 deletion in S. cerevisiae and suggest that Assdr1 plays a crucial role in the regulation of aromatic amino acid metabolism and the synthesis of fermentation flavor compounds. Further investigation into the precise biochemical mechanisms of Assdr1 could contribute to optimizing the metabolic engineering of S. cerevisiae for improved industrial fermentation processes.
3.7. Purification of carbonyl reductases AsSDR1 and AsSDR2
To further characterize the catalytic activity of AsSDR1 and AsSDR2, the enzymes were separately produced using E. coli BL21 and subsequently purified. After 20 h of induction of the corresponding E. coli BL21(DE3) strains HCE010 and HCE020, cells were collected and proteins were purified using Ni–NTA. The apparent molecular weights of AsSDR1 and AsSDR2 deduced from SDS-PAGE gels corresponded to those calculated from the amino acid sequences (27.5 kDa and 28.8 kDa, respectively; Fig. S16). The purity of the purified AsSDR1 and AsSDR2 was estimated to exceed 98 % based on a grayscale analysis of the SDS-PAGE gel after protein staining with Coomassie brilliant blue G250.
3.8. Biochemical properties of purified carbonyl reductase
The effect of temperature and pH on catalytic activity was determined for AsSDR1 and AsSDR2. The activity was tested at different temperatures ranging from 20 to 50 °C. As shown in Fig. 4a, the optimal temperature for AsSDR1 and AsSDR2 reductase activity was 30 °C. The activity was tested at pH values ranging from 3.0 to 9.0. As shown in Fig. 4b, the optimal pH for AsSDR1 and AsSDR2 reductase activity was 8.0; as the pH decreased, enzyme activity gradually decreased.
Fig. 4.
Optimal Reaction Conditions for AsSDR1 and AsSDR2. (a) Purifed enzymes were reacted at different temperatures and assayed at various temperatures from 20 to 50 °C; (b) Purified enzymes were mixed with different buffers and assayed at various pH values, from 3.0 to 10.0. The values provided are the means of three independent replicates.
Enzyme activity units were defined based on wet cell weight, with the reaction system standardized to contain 1.5 g/L substrate and 3 % (v/v) ethanol. Herein, one enzyme activity unit (U) was defined as the amount of enzyme required to generate 1 μmol/L of product per minute under standard conditions (30 °C). The unit activity of AsSDR1(152.40 U/g) was higher than that of AsSDR2(100.39 U/g).
Currently, compared to other studies, the investigation of AsSDR1 in this work remains incomplete and insufficiently comprehensive. This study focuses solely on evaluating the substrate activity of the engineered strain HCE010, hence the decision to measure crude enzyme activity. In contrast, the RasADH literature [11] modified the binding pocket to accommodate bulkier substituted cyclopentanediones and compared the activity of purified enzymes—an aspect not considered in this study regarding extended substrate analogs. Meanwhile, the TbADH patent [12] prioritized enhancing the enzyme's isopropanol tolerance, enabling the use of isopropanol as both a solvent and cosubstrate to improve conversion rates.
This experiment also explored alternative solvents (e.g., methanol, isopropanol) to increase product yield. However, all modified solvent formulations invariably led to statistically significant reductions. It is hypothesized that S. cerevisiae AS2.346 and HCE010 exhibit high ethanol tolerance but are relatively sensitive to other organic solvents (Fig. S17). The AsSDR1 enzyme will be subjected to further modifications in future investigations.
3.9. Selection of carbonyl reductase mutants
Using AutoDock 4.1, ethyl secodione was docked into the substrate binding site of AsSDR1 (Fig. 5a). As shown in Fig. 5b, the hydride of C4 from the nicotinamide ring of NADPH attacked the carbon atom of the carbonyl group, Ser137 stabilized and polarized the carbonyl group of the substrate, and phenol Tyr150 donated a proton to the oxygen atom of the carbonyl group. Lys154 enhanced the acidity of Tyr150 and interacted with the nicotinamide ribose group. In addition, the MOLE Online website was used for substrate binding channel prediction (Fig. 5c). The major sites in the channel included: Leu89, Phe144, Tyr147, Ile180, Met185, Gln186, Ile189, Phe206, and Leu209. Alanine scanning mutagenesis was performed on all the amino acid residues mentioned above (Table 1, Fig. S18). When observing from the entrance of the binding channel towards the binding center, it was showed Leu89, Ile180, and Ile189 obstruct substrate entry from different orientations (Fig. S19). Leu89, Ile180, and Ile189 were selected as target residues for mutagenesis with varying side-chain lengths and polarities to optimize enzymatic activity. The yields of the main products of I180V were almost the same as those in the alanine scanning. However, the yields of their by-products increased to a certain extent (Fig. S20).
Fig. 5.
Molecular docking and mechanism. (a) Docking of ethyl secodione into the substrate binding site of AsSDR1. (b) Schematic view of the AsSDR1 reduction reaction mechanism adapted to ethyl secodione. (c) Substrate binding channel prediction of AsSDR1. (d) Diagram of AsSDR1 protein-substrate binding patterns; (e) Diagram of AsSDR1-L89A protein-substrate binding patterns; (f) Diagram of AsSDR1-I180A protein-substrate binding patterns; (g) Diagram of AsSDR1-I189A protein-substrate binding patterns; (h) Change in the number of protein-substrate hydrogen bonds of AsSDR1; (i) Change in the number of protein-substrate hydrogen bonds of AsSDR1-L89A; (j) Change in the number of protein-substrate hydrogen bonds of AsSDR1-I180A; (k) Change in the number of protein-substrate hydrogen bonds of AsSDR1-I189A; (l) RMSF value of the amino acid residues of the protein.
Compared with wild-type AsSDR1 (HCE010), the yield of HCE011 (single mutant L89A) increased from 71.40 to 81.85 %, the yield of HCE012 (single mutant I180A) increased from 71.40 to 98.26 %, and the yield of HCE013 (single mutant I189A) increased from 71.40 to 97.61 %. HCE014 (double mutant I180A/I189A) showed low enzyme activity (Table 1).
3.10. Molecular docking and molecular dynamics simulation
To analyze the conformational changes of the protein-substrate complexes and the reasons underlying the improved transformation efficiency of the mutants, molecular dynamics simulations were carried out based on the docking conformation. The RMSD value of the protein was calculated based on the crystal structure. In the last 10 ns or more, the amplitude of the oscillation of the RMSD value was less than 0.5 Å, indicating that the system was in equilibrium (Fig. S21a).
After the protein-substrate complex reached equilibrium, the MD attack distance for the first 50 ns was analyzed. The experimental results showed that the average attack distances of wild-type AsSDR1, L89A, I180A and I189A were 3.60, 3.41, 3.48 and 3.06 Å, respectively (Fig. S21b). Compared with the wildtype, the three mutants had shorter effective attack distances between NADPH-H4N and substrate C17 to a certain extent, which improved the transformation efficiency (Fig. 5d–g).
In addition, the stability of the amino acid residues and the number of hydrogen bonds formed were also key factors in determining the conversion efficiency.
The number of hydrogen bonds bound to the substrate by the three mutants was greater than that of wild-type AsSDR1 bound to the substrate, stabilizing the structure of the protein-substrate complex so that it was more conducive to a reaction, thus improving the transformation efficiency (Fig. 5h–k).
The RMSF values of the three mutants were not significantly different from those of the wild type, indicating that the mutations at these three loci had little effect on the structure of the active site pocket. However, at the three mutation sites, the shift of the residues was significantly smaller, indicating that this region was conformationally more stable compared to the wild type (Fig. 5l).
Substrate entry channels for AsSDR1 and its mutants were analyzed using CaverWeb (https://loschmidt.chemi.muni.cz/caverweb/). The amino acid at position 89 controls the size of the active cavity inside the substrate and NADPH binding, and amino acids at positions 180 and 189 control the two directions at the entrance of the substrate binding channel (Fig. S19). Therefore, single-point mutations of these three residues effectively increased the yield.
When AsSDR1 underwent a two-point mutation, although the minimum radius of the channel did not expand significantly, the inlet and inner size of the active cavity changed at the same time. This produced an oversized active center, which led to off-target substrate binding and reduced conversion.
3.11. Overexpression of the AsSDR1 mutant in HCY000
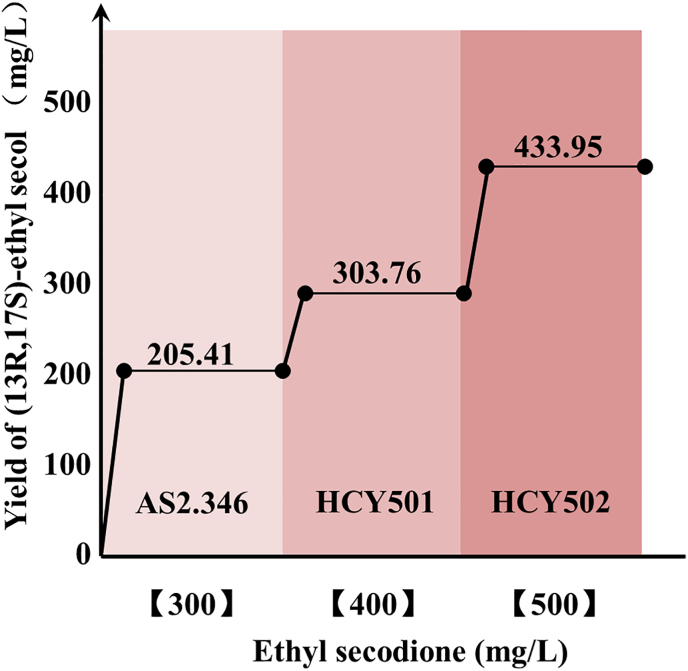
With the goal to increase productivity, the mutant gene encoding Assdr1-I180A was overexpressed under the control of the strong constitutive promoter TDH3 in HCY000. Gene overexpression was confirmed by qPCR (2.5-fold increased expression on average as compared to the wild-type strain, Fig. S22a). The specific growth rates of HCY501 were not significantly different than that of the parental strain (Fig. S23). As described in the method, the biotransformation reaction was carried out. The substrate concentration was 6.6 g/L. Compared with wild-type HCY000, the product yield of HCY501 increased 47.88 % (Fig. 6).
Fig. 6.
Improvement of the yield of (13R,17S)-ethyl secol by S.cerevisiae strains. The x-axis values indicate the ethyl secodione dosage.
3.12. Engineering of cofactor metabolism
As demonstrated above, the reduction of ethyl secodione by AsSDR1 requires NADPH as a cofactor. Therefore, rapidly regenerating this cofactor from NADP+ is a key factor for increasing (13R,17S)-ethyl secol productivity in strain HCY501. The NADH kinase in mitochondria generates NADPH from NADP+. In yeast, the enzyme is encoded by gene pos5 and its first 17 amino acids form a mitochondrial targeting peptide. Therefore, overexpressing the truncated pos5Δ17 in HCY501 regenerates the NADPH consumed by the transformation reaction. Gene overexpression in the transformed strain was confirmed by qPCR (1.68-fold increased expression on average as compared to the wild-type strain; Fig. S22b). The growth rate of HCY502 was significantly accelerated (Fig. S23). The biotransformation reaction was then performed as described in the Methods section. With a substrate concentration of 8.3 g/L, compared with wild-type HCY000, the product yield of HCY502 increased 111.25 % (Fig. 6).
Wang et al. [20] employed wet cells of Geotrichum candidum ZJPH1704 as the enzyme source (300 g/L based on buffer volume) for substrate reduction. In the absence of the co-substrate glycerol, when the substrate concentration was 7.5 g/L (buffer volume basis) with 7 % ethanol added as cosolvent, the product yield reached 20.5 % with 100 % e.e., achieving a space-time yield of 7.12 mg/(L·h) per gram of wet cells. The space-time yields for strains AS2.346 and HCY502 were 4.28 mg/(L·h) and 9.04 mg/(L·h), respectively. Studies have demonstrated that the catalytic activity of Saccharomyces cerevisiae HCY502 surpasses that of G. candidum ZJPH1704. Furthermore, the production process involving G. candidum ZJPH1704 requires additional centrifugation and resuspension steps prior to substrate feeding, which consequently increases production costs.
Overexpression of key enzymes or increasing the copy number of key enzymes in engineered strains is a common means of increasing yield in synthetic biology. Cheng et al. [16] isolated two erythritose reductase genes (ER10 and ER25) in Yarrowia lipolytica and overexpressed them, producing an average yield increase of 14.7 % and a maximum yield increase of 31.2 %.
Increasing the supply of key enzyme cofactors is also a common means of increasing yield in metabolic engineering. Zhao et al. [21] overexpressed glucose-6-phosphate dehydrogenase ZWF1 and NAD kinase POS5 in Saccharomyces cerevisiae, which ultimately increased the yield of β-carotene by 18.8 and 65.6 %, respectively. Liu et al. [22] performed site-directed mutagenesis of the zwf and gnd genes in the pentose phosphate pathway in Corynebacterium glutamicum, which alleviated feedback inhibition and increased the NADPH supply by 151.8 %. Subsequently, a series of metabolic engineering operations were performed on the strain that increased the supply of NADPH by 348.2 % and the yield of l-methionine by 64.1 %.
Increasing the expression of steroid transporters on cell membranes is also an important means of increasing the yield of biotransformations. Chen et al. [23] integrated ClCDR4, an ABC transporter, into Saccharomyces cerevisiae. The engineered strain completely transported acetylated corticosterone into cells within 24 h, thereby increasing the yield of hydrocortisone to 268 mg/(L⋅d). Xu et al. [24] identified PDR11 as the main transporter of rubusoside by knocking out eight PDR subfamily proteins individually. By overexpressing PDR11, the yield of rubusoside increased to 129.8 % of that of the control strain, reaching 155.6 mg/L. In the future, the ABC transporter gene that encodes sterol transport will be knocked out to determine the protein that transports ethyl secodione. Subsequently, the transformation efficiency will be improved by increasing the transport efficiency of the substrate.
4. Conclusion
We isolated and characterized the carbonyl reductase enzyme of Saccharomyces cerevisiae AS2.346. Both AsSDR1 and AsSDR2 were able to reduce ethyl secodione to (13R,17S)-ethyl secol in vitro. Due to differences in subcellular localization, Assdr1 was identified as the functional gene in Saccharomyces cerevisiae AS2.346. Metabolomics analysis revealed that its function was similar to that of sepiapterin reductase. Through protein engineering, it was found that single-point mutations L89A, I180A, and I189A increased the activity of AsSDR1. Overexpression of the gene assdr1-I180A, together with engineering to increase NADPH metabolism, significantly increased (13R,17S)-ethyl secol production.
CRediT authorship contribution statement
Qixin Liang: Writing – original draft, Methodology, Investigation. Wanqing Guo: Methodology, Investigation. Xiaoshuang Sun: Methodology, Investigation. Xianpu Ni: Supervision. Huanzhang Xia: Writing – review & editing, Supervision.
Data statement
The datasets supporting the conclusions of this article are included within the article and its additional files.
Funding
This work was supported by the grants from the Natural Science Foundation of Liaoning (2023-MS-199).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank LetPub (www.letpub.com.cn) for its linguistic assistance during the preparation of this manuscript.
Footnotes
Peer review under the responsibility of Editorial Board of Synthetic and Systems Biotechnology.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2025.04.016.
Contributor Information
Xianpu Ni, Email: nixianpu126@126.com.
Huanzhang Xia, Email: xiahz612@sina.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Tang R., Ren X., Xia M., Shen Y., Tu L., Luo J., et al. Efficient one-step biocatalytic multienzyme cascade strategy for direct conversion of phytosterol to C-17-Hydroxylated steroids. Appl Environ Microbiol. 2021;87 doi: 10.1128/aem.00321-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kristan K., Stojan J., Adamski J., Lanisnik Rizner T. Rational design of novel mutants of fungal 17beta-hydroxysteroid dehydrogenase. J Biotechnol. 2007;129:123–130. doi: 10.1016/j.jbiotec.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y., Wang Y., Chen X., Feng J., Wang M., Wu Q., et al. Modulating the active site lid of an alcohol dehydrogenase from Ralstonia sp. enabled efficient stereospecific synthesis of 17β-hydroxysteroids. Enzym Microb Technol. 2021;149 doi: 10.1016/j.enzmictec.2021.109837. [DOI] [PubMed] [Google Scholar]
- 4.Shen J., Che Y., Showell E., Chen K., Cheng L. Interventions for emergency contraception. Cochrane Database Syst Rev. 2019;1:Cd001324. doi: 10.1002/14651858.CD001324.pub6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gemzell-Danielsson K., Marions L. Mechanisms of action of mifepristone and levonorgestrel when used for emergency contraception. Hum Reprod Update. 2004;10:341–348. doi: 10.1093/humupd/dmh027. [DOI] [PubMed] [Google Scholar]
- 6.Lalitkumar P.G., Berger C., Gemzell-Danielsson K. Emergency contraception. Best Pract Res Clin Endocrinol Metabol. 2013;27:91–101. doi: 10.1016/j.beem.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Han G., Ma Z., Dai Y. Development of total synthesis of coutaceptive steroid drugs in China. Chin J Pharm. 2000;31:231–235. [Google Scholar]
- 8.Steroid Hormone Group Shanghai Institute of organic Chemistry, Chinese Academy of Sciences. Total synthesis of the oral contraceptives D-norgestrel and D-18-methyl-norgestrienone. Chemistry. 1976:23–25. doi: 10.14159/j.cnki.0441-3776.1976.05.006. [DOI] [Google Scholar]
- 9.Fa Y., Xu S., Ma S., Zhang L. Studies on the microbiological preparation of 13β ethyl-3-methoxy-8,14-seco-1,3,5(10),9(11)-estratetraene-17β-ol,14-one. Acta Microbiol Sin. 1978;18:232–238. doi: 10.13343/j.cnki.wsxb.1978.03.010. [DOI] [Google Scholar]
- 10.Contente M.L., Molinari F., Serra I., Pinto A., Romano D. Wiley Online Library; 2016. Stereoselective enzymatic reduction of ethyl secodione: preparation of a key intermediate for the total synthesis of steroids. [Google Scholar]
- 11.Chen X., Zhang H., Maria-Solano M.A., Liu W., Li J., Feng J., et al. Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase. Nat Catal. 2019;2:931–941. doi: 10.1038/s41929-019-0347-y. [DOI] [Google Scholar]
- 12.Chen X., Zhang H., Liu X., Feng J., Wu Q., Zhu D., et al. 2021. Carbonyl reductase mutant and application thereof. [Google Scholar]
- 13.Sambrook J., Russell D.W. third ed. Cold Spring Harbor Laboratory Press; New York: 2001. Molecular cloning: a laboratory manual. [Google Scholar]
- 14.Sherman F., Fink G., Hicks J. Cold Spring Harbor Laboratory Press; 1987. Methods in yeast genetics: a laboratory course manual. [Google Scholar]
- 15.Ryan O.W., Poddar S., Cate J.H. CRISPR-Cas9 genome engineering in Saccharomyces cerevisiae cells. Cold Spring Harb Protoc. 2016;2016 doi: 10.1101/pdb.prot086827. [DOI] [PubMed] [Google Scholar]
- 16.Cheng H., Wang S., Bilal M., Ge X., Zhang C., Fickers P., et al. Identification, characterization of two NADPH-dependent erythrose reductases in the yeast Yarrowia lipolytica and improvement of erythritol productivity using metabolic engineering. Microb Cell Fact. 2018;17:133. doi: 10.1186/s12934-018-0982-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J.J., Zhang G.C., Kwak S., Oh E.J., Yun E.J., Chomvong K., et al. Overcoming the thermodynamic equilibrium of an isomerization reaction through oxidoreductive reactions for biotransformation. Nat Commun. 2019;10:1356. doi: 10.1038/s41467-019-09288-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Christiano R., Nagaraj N., Fröhlich F., Walther T.C. Global proteome turnover analyses of the Yeasts S. cerevisiae and S. pombe. Cell Rep. 2014;9:1959–1965. doi: 10.1016/j.celrep.2014.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P., Xiao M., Chen H., Miao Y. 2020. A biological preparation method for a key chiral intermediate of levonorgestrel. CN107881202B. [Google Scholar]
- 21.Zhao X., Shi F., Zhan W. Overexpression of ZWF1 and POS5 improves carotenoid biosynthesis in recombinant Saccharomyces cerevisiae. Lett Appl Microbiol. 2015;61:354–360. doi: 10.1111/lam.12463. [DOI] [PubMed] [Google Scholar]
- 22.Liu B., Sun X., Liu Y., Yang M., Wang L., Li Y., et al. Increased NADPH supply enhances glycolysis metabolic flux and L-methionine production in Corynebacterium glutamicum. Foods. 2022;11 doi: 10.3390/foods11071031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J., Fan F., Qu G., Tang J., Xi Y., Bi C., et al. Identification of Absidia orchidis steroid 11β-hydroxylation system and its application in engineering Saccharomyces cerevisiae for one-step biotransformation to produce hydrocortisone. Metab Eng. 2020;57:31–42. doi: 10.1016/j.ymben.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 24.Xu Y., Wang X., Zhang C., Zhou X., Xu X., Han L., et al. De novo biosynthesis of rubusoside and rebaudiosides in engineered yeasts. Nat Commun. 2022;13:3040. doi: 10.1038/s41467-022-30826-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.