Abstract
We have previously described a 160-bp enhancer (BCE-1) in the bovine β-casein gene that is activated in the presence of prolactin and extracellular matrix (ECM). Here we report the characterization of the enhancer by deletion and site-directed mutagenesis, electrophoretic mobility shift analysis, and in vivo footprinting. Two essential regions were identified by analysis of mutant constructions: one binds C/EBP-β and the other binds MGF/STAT5 and an as-yet-unidentified binding protein. However, no qualitative or quantitative differences in the binding of these proteins were observed in electrophoretic mobility shift analysis using nuclear extracts derived from cells cultured in the presence or absence of ECM with or without prolactin, indicating that prolactin- and ECM-induced transcription was not dependent on the availability of these factors in the functional cell lines employed. An in vivo footprinting analysis of the factors bound to nuclear chromatin in the presence or absence of ECM and/or prolactin found no differences in the binding of C/EBP-β but did not provide definitive results for the other factors. Neither ECM nor prolactin activated BCE-1 in transient transfections, suggesting that the chromosomal structure of the integrated template may be required for ECM-induced transcription. Further evidence is that treatment of cells with inhibitors of histone deacetylase was sufficient to induce transcription of integrated BCE-1 in the absence of ECM. Together, these results suggest that the ECM induces a complex interaction between the enhancer-bound transcription factors, the basal transcriptional machinery, and a chromosomally integrated template responsive to the acetylation state of the histones.
It is now well established that the processes of development and differentiation depend on a cell’s ability to correctly perceive its microenvironment (reviewed in references 1 and 43). A key component of this environment is the extracellular matrix (ECM). The ECM is an organized network of glycoproteins, proteoglycans, and glycosaminoglycans, components important for cell morphology as well as for signal transduction via cell surface integrins and ultimately for tissue-specific gene expression (reviewed in reference 43).
The mammary gland appears to be particularly well suited for the study of ECM-induced differentiation and gene expression. In the adult animal, the gland develops after puberty and functionally differentiates in response to pregnancy. The mechanisms involved in these developmental processes are complex and guided by various hormones (54), growth factors (53), and the ECM (3). Milk protein expression is initiated at mid-pregnancy and correlates with the synthesis and deposition of a specialized laminin-rich ECM during alveolar development. Expression of these milk proteins can be used as markers for the differentiated state of the gland. In the last decade, a number of model systems using mammary epithelial cells to study ECM-dependent gene regulation have been developed. These range from primary cultures to cloned cell lines which undergo a three-dimensional reorganization in gelatinous matrices to form alveolus-like structures capable of synthesizing and vectorially secreting milk proteins, analogous to their in vivo counterparts in the lactating mammary gland (references 2 and 30 and references therein).
Studies with SCP2 (11) and CID-9 (44) cell lines derived from the COMMA 1D cell strain (8), itself derived from the mammary tissue of midpregnancy mice, have shown that induction of endogenous β-casein requires both an ECM-induced change in cell shape and a β1-integrin-mediated biochemical signaling by laminin, a major component of mammary basement membrane (42, 52). Downstream nuclear events associated with this integrin signal transduction pathway have been analyzed with stable transfectants of CID-9 cells with the bovine β-casein promoter linked to the chloramphenicol acetyltransferase (CAT) reporter gene. These studies clearly demonstrated that the transcriptional regulation of this gene is dependent on the presence of both ECM and lactogenic hormones (44). Deletion analysis of this promoter identified a 160-bp transcriptional enhancer (BCE-1) capable of conferring ECM and hormonal regulation in either orientation to the inactive proximal β-casein promoter (−121 to +42) (46).
Many cis-acting transcription elements have been identified in the rat and mouse β-casein genes. Their functional role in induction of transcription has been studied in mammary glands of transgenic animals, in primary mammary epithelial cells, and in other cell lines such as HC11 (also derived from COMMA 1D) (14, 19, 29, 63). These studies have identified three major trans-acting factors that are involved in the hormone-induced transcriptional activation of β-casein. Prolactin has been shown to activate transcription via STAT5, originally identified as mammary gland factor (MGF) (20, 47, 56, 57–59). STAT5 binding sites are found in many other milk protein promoters, with the STAT binding sites in the sheep β-lactoglobulin gene being particularly well characterized (5, 6, 51, 58). C/EBP binding sites are required for the hormonal induction of β-casein expression (16, 41, 49), and its activity may be influenced by glucocorticoid receptor, which has been shown to enhance prolactin-induced expression of β-casein (15, 25, 41, 44, 50).
In this study, we found that the binding sites for C/EBP and STAT5 are contained within the ECM-responsive element BCE-1. Furthermore, we show that these elements, as well as an unidentified binding protein (UBP) site are critical for ECM and prolactin responsiveness. Unexpectedly, we also find that despite the dramatic increase in transcription when the cells are cultured on ECM, these factors appear to be bound to BCE-1 even when cells are cultured on plastic in the absence of prolactin. Based on the induction of BCE-1 transcriptional activity in cells cultured on plastic after treatment with histone deacetylase inhibitors, we propose that the ECM may allow transcription of the β-casein gene via a mechanism that involves modulation of histone acetylation.
MATERIALS AND METHODS
Plasmids. (i) Cloning of the deletions.
The first 100 bp and the 3′ 61 bp of BCE-1 were isolated by PCR from the expression plasmid BBC (termed BCE-1/ER-1/CAT in reference 46). These fragments were cloned into ER-1 and checked by sequencing.
(ii) Site-specific mutations.
Site-specific mutations were introduced into BBC with a mutagenesis kit (transformer site-directed mutagenesis kit; Clontech, Palo Alto, Calif.). The 30-bp primers for mutagenesis included 10 bp of BCE-1 sequence flanking the mutation sequence GCTCTAGAGC (an XbaI site [underlined] flanked by GCs). The correct introduction of each mutation was confirmed by sequencing.
(iii) Plasmids for transient transfections.
MMTV/CAT, MMTV/β-cas/CAT, and SV40/β-cas/CAT were described previously (45); LTR/Luc is described in reference 31.
Cell passage and differentiation.
CID-9 cells (44) and their transfected derivatives were passaged in Dulbecco modified Eagle medium (DMEM)–F-12–5% fetal calf serum–insulin (5 μg/ml) (referred to as growth medium) and induced to differentiate in DMEM–F-12–insulin (I) (5 μg/ml) without or with 1 μg of hydrocortisone (H) per ml and/or 3 μg of prolactin per ml (P) as described previously (differentiation medium [46]). Sodium butyrate (Sigma Chemical Co., St. Louis, Mo.) and trichostatin A (Wako Pure Chemical Industry, Richmond, Va.) were prepared as 1,000× stock solutions in water and ethanol, respectively. In experiments where sodium butyrate and trichostatin A were used, the cells were treated 48 h after plating and harvested after 18 h of treatment. In some of the experiments, Matrigel (Collaborative Biomedical Products, Bedford, Mass.), insulin (Gibco/BRL, Bethesda, Md.), and prolactin (Sigma) were used. Stable transfections, cell harvest, and CAT assays were performed as described previously (46).
Transient transfections.
CID-9 cells were plated in growth medium at a density of 3 × 105 per 60-mm-diameter tissue culture plastic dish or at 1.2 × 106 per 60-mm dish in dishes coated with 0.4 mg of polyhema (Sigma [42]) 1 day prior to transfection. Cells were given DMEM–5% fetal calf serum and 5 μg of insulin per ml at least 3 h before transfection. Ten micrograms of test plasmid and 1 μg of RSV/β gal were cotransfected by the calcium phosphate method (46) with the exception that the precipitates were left on the cells for 18 h. The cells were then washed three times with DMEM–F-12 and placed in differentiation medium (IHP). Cells cultured on polyhema were given differentiation medium containing 2% Matrigel as described previously (42). They were harvested 48 h later with Dispase (44). Lysates were assayed for β-galactosidase (β-Gal) activity (Glacto-Light; Tropix, Bedford, Mass.), protein content (Bio-Rad protein assay; Bio-Rad, Hercules, Calif.), and CAT (44) or luciferase activity (12). The thin-layer chromatography (TLC) plates from the CAT assays were exposed to PhosphorImager screens and analyzed with a Molecular Dynamics PhosphorImager. The CAT activity was analyzed and expressed relative to β-Gal activity.
Preparation of nuclear extracts.
Nuclear proteins for electrophoretic mobility shift analysis (EMSA) were extracted by two different procedures (10, 48) with similar results in the quality and quantity of the isolated proteins. Extracts were isolated in the presence or absence of the phosphatase inhibitors sodium fluoride, sodium molybdate, and orthovanadate. Cells cultured on ECM were released by Dispase treatment or harvested by matrix dissolution with phosphate-buffered saline (PBS) (without calcium and magnesium), 5 mM EDTA, and protease inhibitors (0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol, 2 mM benzamidine, 5 μg each of pepstatin and leupeptin per ml) (a kind gift of Steve Farmer, Boston University) or harvested by scraping cells together with the ECM (isolation conditions were as noted in the figure legends). All extracts were quantitated for protein (Bio-Rad), and aliquots were run on sodium dodecyl sulfate (SDS)–12% polyacrylamide gel electrophoresis and visualized by Coomassie blue staining to assess the quality and to independently quantitate the extracts. With the exception of slight ECM contamination in extracts isolated from cells which did not have the ECM removed by Dispase or PBS dissolution, the methods produced similar results in all BCE-1 EMSAs. However, Dispase-treated nuclear extracts displayed an altered NF-1 mobility shift (34a), indicating that these preparative methods may not be equivalent for all nuclear proteins.
EMSA.
Mobility shift probes were labeled as single-stranded oligonucleotides with T4 polynucleotide kinase in the presence of [γ-32P]ATP (>5,000 Ci/mmol); complementary strands were annealed, and the double-stranded probe was gel purified. The amount of labeled probe ranged from 10 to 60 fmol per binding assay. The sequences below represent the coding strands for the following oligonucleotides: A, 5′-TGTATTCCTTTTCTCAGAAATC; B, 5′-ATCACACTTTTTTGCCTGTGGC; D, 5′-CTGTTTATTGCACAATATGT; C/EBP, 5′-TGCAGATTGCGCAATCTGCA (7); and MGF, 5′-GGACTTCTTGGAATTAAGGGA (adapted from reference 47).
Nuclear proteins (10 μg) were incubated for 20 min in binding buffer (20 mM HEPES [pH 7.8], 0.5 mM MgCl2, 1 mM dithiothreitol, 0.5 mM EDTA, 5% glycerol, 80 mM NaCl) with 0.5 μg of poly(dI-dC) and 10,000 cpm of probe in a 20-μl volume. Binding reactions were run on a 5% nondenaturing polyacrylamide gel in 0.25× Tris-borate-EDTA. When present, competitors were double stranded and used at a 50-fold molar excess over the labeled probe. For supershift analysis, antibodies against C/EBP-α, -β, and δ and CRP1 were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.), and those against STAT5a and STAT5b (a kind gift of Lothar Henninghausen, National Institutes of Health) and STAT1 and STAT3 were obtained from Transduction Laboratories Inc. (Lexington, Ky.). The binding reactions were carried out for 20 min at room temperature, and then antibodies were added and the reaction mixtures were set on ice for 30 min before being loaded onto a 4% acrylamide–0.25× Tris-borate-EDTA gel.
Nucleus isolation and exonuclease assay.
Nuclei were isolated from cells as described previously (39), with the exception that cells cultured on ECM were first released by the PBS treatment described above. The digestion of nuclei for exonuclease assays was similar to that of Pennie et al. (39). An aliquot of each nucleus preparation was quantitated for DNA content. Nuclei were thawed on ice and diluted into Workman and Langmore buffer (50 mM NaCl, 50 mM Tris-Cl [pH 8], 0.5 mM MgCl2, 1 mM beta-mercaptoethanol) such that 200-μl aliquots contained 20 μg of DNA. The nuclease digestions were conducted for 15 min at 37°C with the following nuclease concentrations (per microgram of DNA): HinfI, NcoI, and BamHI, 20 U; lambda exonuclease, 1 U; and T7 exonuclease, 50 U. The reactions were stopped by adding an equal volume of stop solution (0.2 M NaCl, 50 mM Tris-Cl [pH 8], 20 mM EDTA, 2% SDS). Protease K (10 μl of a 10-mg/ml solution) was added, and the samples were incubated at 37°C overnight. The samples were then extracted with phenol-chloroform and precipitated with 0.25 M NaCl and isopropanol. Washed pellets were dried and resuspended in water prior to a secondary restriction digestion, which was performed with the following enzymes: StyI or EcoRI for HinfI, BstBI for NcoI, and BsRI for BamHI (2 to 4 U/μg of DNA). Samples were extracted and precipitated as described above. Ten-microgram portions of each sample were subjected to linear amplification with [γ-32P]ATP-labeled oligonucleotides and Taq polymerase. Samples were extracted as described above and precipitated with 0.25 M NaCl and ethanol, resuspended in 80% formamide solution containing bromophenol blue and xylene cyanol, and separated on an 8% denaturing polyacrylamide gel. Dried gels were exposed to a PhosphorImager screen and analyzed with a Molecular Dynamics PhosphorImager. Twenty nanograms of BBC plasmid DNA was subjected to the same treatment described above as a control for primary enzyme cutting and primer extension in the PCR step. Genomic DNA isolated from stably BBC-transfected CID-9 cells was used as a control to demonstrate the ability of lambda and T7 exonuclease to progress through the genomic sequence of BBC. The digestion with the secondary enzyme allows for visualization of the quantity of DNA present in each condition.
RESULTS
Functional elements within BCE-1.
A deletion analysis was first conducted to characterize the functionally important regions within the 160-bp BCE-1 enhancer. Two deletion constructs were made, one containing 100 bp of 5′ end and one containing 60 bp of the 3′ end of the BCE-1 enhancer linked to the transcriptionally inactive −121 to +42 sequence of the bovine β-casein promoter (Fig. 1A). The constructs were stably transfected into CID-9 cells, and the activity of each was compared to that of stably integrated full-length BCE-1. Neither deletion construct was active, in either the presence or absence of ECM (Fig. 1A and data not shown). Therefore, BCE-1 requires the presence of both halves for transcriptional activity. A more detailed mutational analysis was then conducted with linker mutant constructs. Mutations were generated across BCE-1 by the replacement of sequential 10-bp stretches of DNA with an XbaI linker. These constructs were stably transfected into CID-9 cells, and transcriptional activity was assessed in the presence or absence of ECM and/or lactogenic hormones. In the presence of ECM and prolactin five mutations resulted in a loss of at least 90% activity compared to wild-type BCE-1 (Fig. 1B). These mutations were localized to two separate regions within BCE-1. Mu3 and Mu4 lay within the first 100 bp of BCE-1 and span a putative C/EBP binding site (region I). Mu11, Mu12, and Mu13 were in the last 60 bp and include a putative STAT5 binding site and another potential protein binding site (region II) (Fig. 1C). All other mutant constructs had between 30 and 80% of wild-type activity. As found with wild-type BCE-1, none of the mutants activated transcription in the absence of ECM and/or lactogenic hormones (data not shown).
FIG. 1.
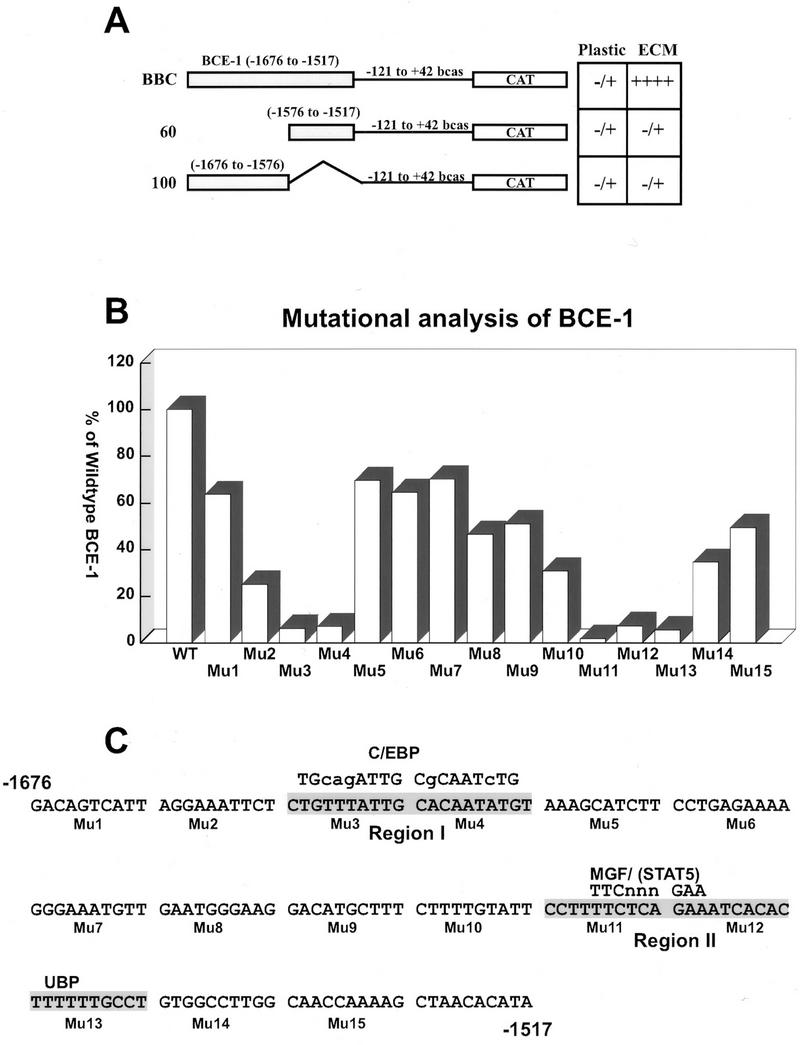
Identification of two functional regions of BCE-1. (A) Schematic representation of full-length BCE-1 (BBC) and two deletion constructs (60 and 100) linked to the minimal bovine β-casein promoter. Sequences are numbered relative to the transcription start site of the endogenous gene. The chart on the right summarizes the transcription activity of the three constructs stably transfected into CID-9 cells. The cells were cultured under differentiation conditions (IHP) without (plastic) or with ECM for 6 days and assayed for CAT activity. −, no detectable activity; +, increasing amounts of transcriptional activity. (B) Transcriptional activity of 15 mutant constructions spanning the entire wild-type BCE-1 enhancer stably transfected into CID-9 cells. Each mutation was generated by replacement of 10 bp of BCE-1 sequence with GCTCTAGAGC (an XbaI site [underlined] flanked by GCs) as described in Materials and Methods. Cells were cultured in IHP on ECM for 6 days and assayed for CAT activity. The graph shows the average of at least three separate transfections for each mutation (Mu1 to Mu15) and shows each mutant’s activity as a percentage of the wild-type BCE-1 (WT) transfected and differentiated at the same time as the mutants. (C) Sequence of the wild-type BCE-1 enhancer and the locations of mutations (Mu1 to Mu15). Regions I and II show the sequence in which mutations knocked out most of the transcriptional activity. The consensus sequence for putative binding sites of C/EBP and STAT are given (uppercase letters denote exact base pair matches). UBP represents a potential protein binding site.
Identification of BCE-1 binding activities by EMSA (i) C/EBP-β binds to region I.
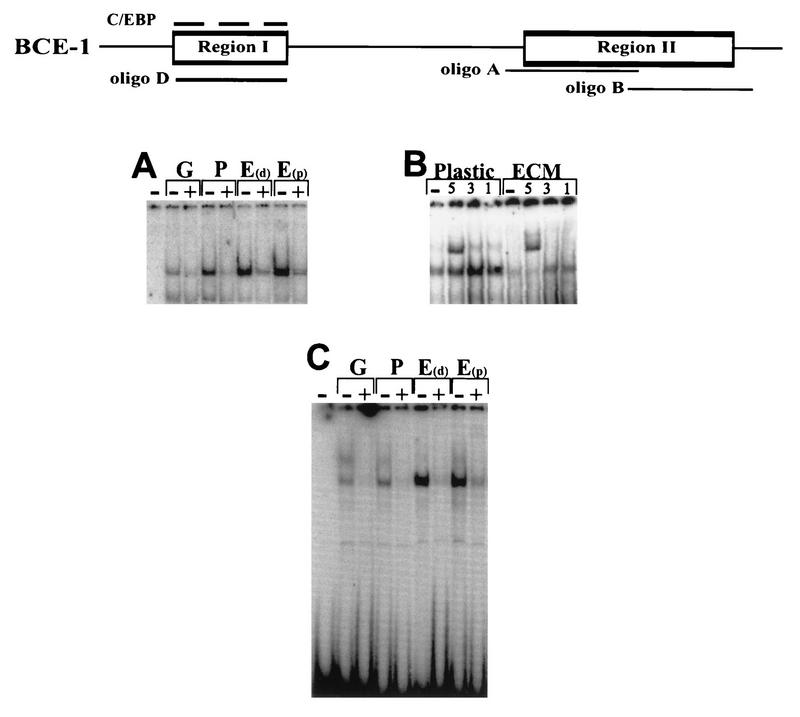
Nuclear extracts from exponentially growing CID-9 cells or CID-9 cells cultured in the presence or absence of ECM and/or lactogenic hormones formed specific complexes with a 20-bp oligonucleotide (oligonucleotide D) that spanned region I, which contains the putative C/EBP binding site. These complexes were competed by an excess of either cold oligonucleotide D (Fig. 2A, “+” lanes; Fig. 2B, lane 4) or a consensus oligonucleotide to C/EBP (Fig. 2B, lanes 5 and 6). There was no difference in the ability to form a complex whether extracts were isolated from (i) actively dividing cells in the presence of serum and the absence of lactogenic hormones (growth), (ii) growth-arrested cells in the absence of serum and the presence of lactogenic hormones (plastic), or (iii) growth-arrested cells in the presence of lactogenic hormones and exogenous basement membrane (i.e., the ECM). The region I complex was supershifted upon incubation with an antibody to C/EBP-β (Fig. 2C, lane 4) but not by an antibody to C/EBPα, C/EBPδ, or CRPI (a C/EBP homolog) (Fig. 2C, lanes 3, 5, and 6). Nuclear extracts from the liver, which is known to be an abundant source of C/EBP-α and -β, formed specific complexes with oligonucleotide D (Fig. 2B, lanes 8 to 10) which could be supershifted by both the C/EBP-α and -β antibodies (Fig. 2C, lanes 9 and 10).
FIG. 2.
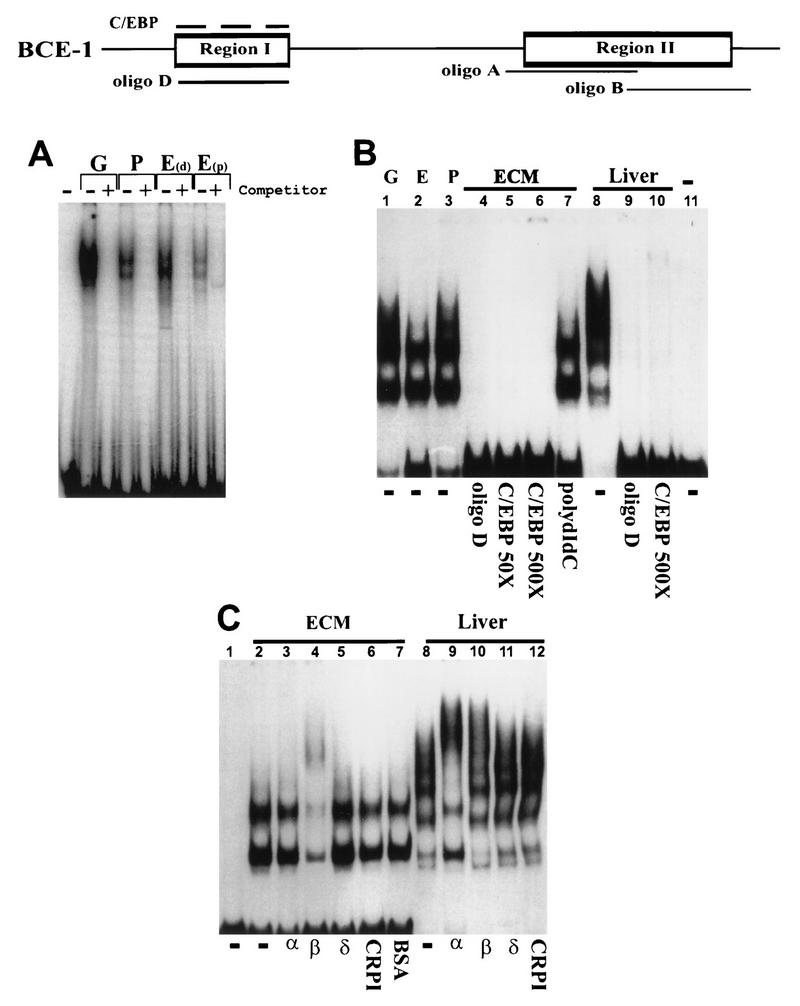
Characterization of the binding activities of proteins interacting with regions I. A schematic of BCE-1 showing the relative positions of oligonucleotides A, B, and D is shown at the top. The C/EBP consensus oligonucleotide is represented by a dashed line above region I. The nuclear extracts isolated from CID-9 cells are designated as follows: G, actively growing cells; P, cells cultured in differentiation medium (IHP); E, cells cultured on ECM in differentiation medium. E(d) cells were harvested with Dispase; E(p) cells were harvested by PBS dissolution. Extracts in A, B, and C were isolated as described previously (references 10 and 48, respectively). Liver extracts were isolated from mouse liver as described previously (48). (A) EMSA using oligonucleotide D. The “+” indicates that a 50× molar excess of unlabeled double-stranded oligonucleotide D was added to the binding mixture. The first lane is labeled probe without extract. (B) EMSA comparing the binding activity of CID-9 cell extracts to that of mouse liver. The competitions are shown below the gel as follows: −, no competitor; oligonucleotide D (50×); consensus C/EBP (50× and 500×), 50× poly(dI-dC) (nonspecific competitor). The last lane is labeled probe without extract. (C) EMSA using oligonucleotide D with CID-9 extracts from cells cultured on ECM in IHP and extracts from mouse liver. The first lane is labeled probe with no extract; α, β, δ, and CRPI represent binding reactions incubated with antibodies to C/EBP-α, -β, and -δ and to CRPI (as described in Materials and Methods). The CID-9 cell extracts are supershifted only with an antibody to C/EBP-β, and the liver extracts are supershifted with antibodies to C/EBP-α and -β.
(ii) Region II binds STAT protein and an additional unidentified binding protein (UBP).
An oligonucleotide that spans the putative STAT5 binding site in BCE-1 (oligonucleotide A) (Fig. 3A) formed specific complexes with either actively growing CID-9 nuclear extracts or CID-9 cells cultured in the presence or absence of ECM. These complexes were competed by an excess of unlabeled oligonucleotide A (Fig. 3A, “+” lanes) or an oligonucleotide to the rat MGF/STAT5 binding site but not by an oligonucleotide to region II which lies outside the STAT site (oligonucleotide B) (data not shown). Supershift analysis of oligonucleotide A with antibodies to STAT5, STAT1, and STAT3 revealed that STAT5 is present in nuclear extracts from cells cultured on plastic and ECM (Fig. 3B). Regardless of the culture conditions, there were no differences in the binding activity present in the various nuclear extracts. This included cells cultured with or without lactogenic hormones.
FIG. 3.
Characterization of the binding activities of proteins interacting with region II. The schematic and the extracts isolated were as described for Fig. 2. (A) EMSA using oligonucleotide A from region II as a probe with nuclear extracts from CID-9 cells as described for Fig. 2. +, lanes with a 50× molar excess of unlabeled double-stranded oligonucleotide A. (B) EMSA using oligonucleotide A with nuclear extracts from CID-9 cells cultured on plastic in IHP and ECM in IHP. −, no antibody; 5, addition of 1 μl of a 1:10 dilution of a 1:1 mixture of STAT5a and STAT5b antibodies to the binding reaction; 1 and 3, addition of 1 μl of antibody to STAT1 and STAT3, respectively. (C) EMSA using oligonucleotide B from region II as a probe. +, a 50× molar excess of unlabeled double-stranded oligonucleotide B (the quantitative differences in binding activity observed in this gel varied in at least three different EMSAs).
Oligonucleotide B, which spans the 3′ end of region II, also formed a specific complex (Fig. 3C). This complex could be competed by an excess of unlabeled oligonucleotide B (Fig. 3C, “+” lanes) but not by oligonucleotide A or an oligonucleotide that contains the Mu13 mutation (data not shown). Although there are differences in the intensities of some complexes among nuclear extracts from cells cultured under different conditions, these differences were not consistent from assay to assay with either the same or independently isolated nuclear extract preparations. Therefore, no definitive conclusions can be drawn by the relative intensities. However, the binding activities and their sensitivity to competition were always consistent among extracts and different preparations.
The ability of nuclear proteins to bind to oligonucleotides in in vitro binding assays does not guarantee that these same proteins will have the access and/or the ability to bind to a chromatin template. We therefore examined whether the factors were differentially bound on the intact chromatin template in cells cultured with or without ECM and/or prolactin.
The ECM does not appear to restrict or induce factor loading onto the chromatin template.
In order to determine whether ECM-dependent transcription results from changes in factor access to the chromatin template, we conducted in vivo exonuclease footprinting analyses. We examined factors bound to chromatin in nuclei isolated from BBC-transfected CID-9 cells which were cultured with or without ECM and/or prolactin, leading to conditions of active and inactive transcription, respectively. To ascertain the transcriptional state of the cells, we routinely performed CAT assays using an aliquot of cells taken before the nuclei were isolated (Fig. 4A). To analyze region I (Fig. 4C), nuclei were treated with the restriction enzyme NcoI with or without the exonuclease lambda or T7. The exonuclease stops were within the boundaries of the C/EBP and OctI binding sites. However, no differences in the stop patterns were observed under any of the culture conditions tested. A number of control reactions were conducted to confirm the specificity of the exonuclease stops. Genomic DNA isolated from the stably transfected CID-9 cells was used as a control for DNA sequence-induced stops (Fig. 4B). BBC plasmid was used as a control for the ability of the restriction enzyme to cut under the digestion conditions used, as well as a control for possible pausing during the PCR primer extension analysis. In addition, two different exonucleases (lambda and T7) were used, since they have different abilities to penetrate DNA that contains bound protein. The control reactions show that the stops are not due either to sequence-dependent pausing by the exonuclease or to peculiarities specific to one nuclease.
FIG. 4.
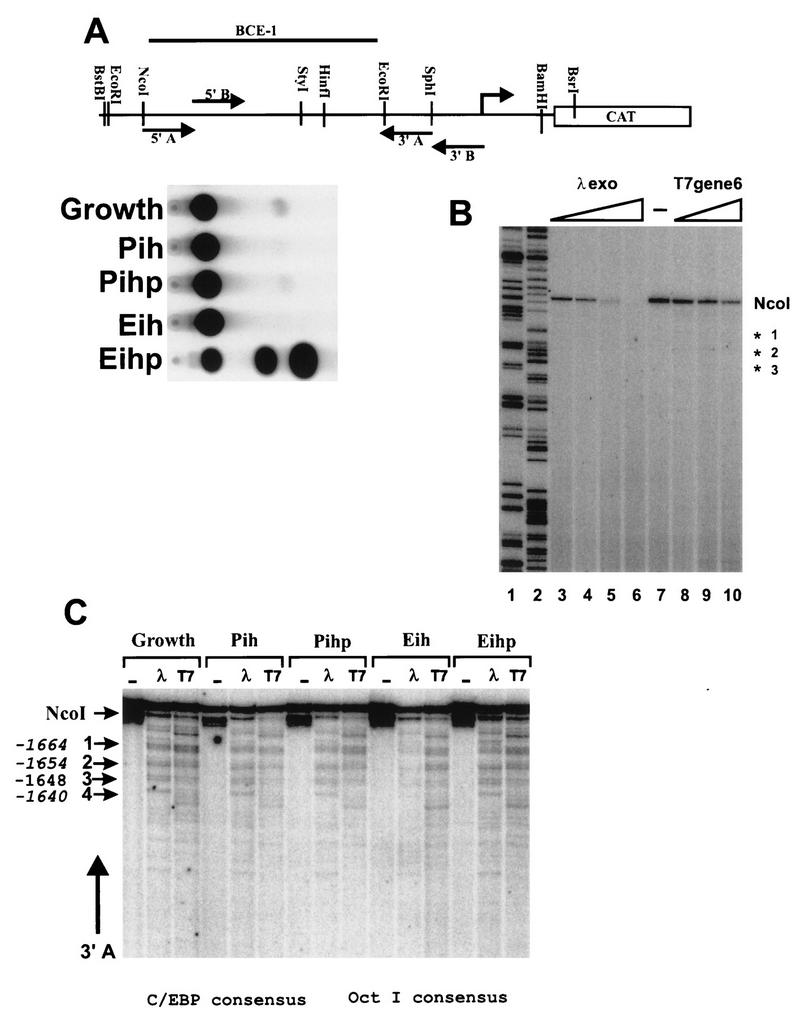
Exonuclease analysis of BCE-1 shows identical stop patterns for both the active and inactive states of the promoter for C/EBP. (A) Diagram of the BBC construct identifying the primary and secondary enzyme digestion sites and the relative location of the primers. Corresponding CAT assays of lysates collected from an aliquot of cells from each culture condition prior to nucleus isolation. The cells cultured on ECM were isolated by PBS dissolution. P, plastic; i, insulin; h, hydrocortisone; p, prolactin; E, ECM. (B) Exonuclease control for genomically induced pausing of the lambda and T7 exonucleases. Genomic DNA isolated from stably BBC-transfected CID-9 cells was cut with NcoI, and aliquots were treated with increasing amounts of exonuclease for 15 min at 37°C. DNA was purified and subjected to primer extension using labeled primer 3A. Lanes: 1 and 2, BBC plasmid DNA extended with labeled primer 3A incorporating ddGTP and ddATP, respectively; 3 to 6, 0.5, 1, 2.5, and 5 U of lambda exonuclease per μg of genomic DNA; 7, control (no exonuclease); 8 to 10, 5, 10, and 25 U of T7 exonuclease per μg of genomic DNA. (C) Exonuclease analysis of region I. NcoI was used to open the chromatin DNA for exonuclease entry. The secondary enzyme was BstBI, and primer extensions were performed with oligonucleotide 3′A. Major stops are designated by arrows 1 to 4, and their positions within the BCE-1 sequence are noted beside the arrows. The positions of the consensus sequences for C/EBP and OctI are noted below the figure.
We have shown by EMSA that OctI forms a specific complex with an oligonucleotide corresponding to the sequence of the wild-type Mu5 region of BCE-1, which can be competed by an excess of unlabeled oligonucleotide as well as by an OctI consensus oligonucleotide (data not shown). However, since there was no significant decrease in transcriptional activity when this site was mutated, we believe that OctI modulation of ECM-dependent transcriptional activation is not absolutely required. The footprinting results were consistent with the OctI site being occupied in vivo.
Region II of BCE-1 was analyzed with the restriction enzyme HinfI to open the chromatin. Exonuclease analysis revealed several strong and weak stops within the STAT and UBP sites. Analysis of control genomic DNA also revealed strong sequence-dependent stops, making it difficult to definitively attribute the weaker stops to chromatin-dependent pausing of the exonuclease. The exonuclease assay seems to specifically detect C/EBP and OctI binding. However, exonuclease stops are present in chromatin from nuclei isolated from actively growing cells (growth) as well as in cells cultured in the presence or absence of ECM and/or prolactin (i.e., IH versus IHP).
Despite the strong transcriptional induction by ECM and prolactin, no differences were observed in the binding of nuclear proteins to the functionally important regions of BCE-1, in vitro. Furthermore, while only C/EBP could definitively be shown to bind in vivo, there were no differences with or without ECM. We therefore wished to determine whether ECM and/or prolactin could induce transcription via the BCE-1 enhancer on a nonintegrated template in a transient-transfection assay.
ECM and prolactin do not induce transcriptional activation of transiently transfected templates.
We examined the transcriptional activity of several chimeric constructs in transient-transfection assays. When integrated into the genome, these constructs respond either positively or negatively to the presence of ECM (37a, 45). In these experiments, the mouse mammary tumor virus (MMTV) and simian virus 40 (SV40) enhancers were linked to the bovine β-casein promoter or the minimal (−110) MMTV promoter (Fig. 5C). The grid shown in Fig. 5C summarizes the results from the transient-transfection assays performed with these constructs, as well as the activities of these same constructs when stably integrated. ECM did not induce BCE-1-dependent transcription on a transient template (no detectable CAT activity), even though endogenous β-casein was differentially expressed under these culture conditions, as shown by the immunoprecipitation of mouse milk proteins from parallel plates of transfected cells (Fig. 5A). To address the question of whether the basal β-casein promoter is capable of supporting transient transcription, the promoter was linked to the SV40 enhancer. This construct had transient transcriptional activity (Fig. 5C). This result clearly demonstrates that the absence of transcription after transient transfection of BCE-1 constructs and treatment of cells with ECM was not due to the failure to utilize the β-casein promoter. The requirement for stable integration of BCE-1 is striking. We therefore decided to examine whether alterations in the chromatin structure of the integrated BCE-1 element could influence transcriptional activity. Because of the strong correlation between histone acetylation and gene activation (33, 55, 60), we analyzed the effect of histone modification on ECM-induced transcription.
FIG. 5.
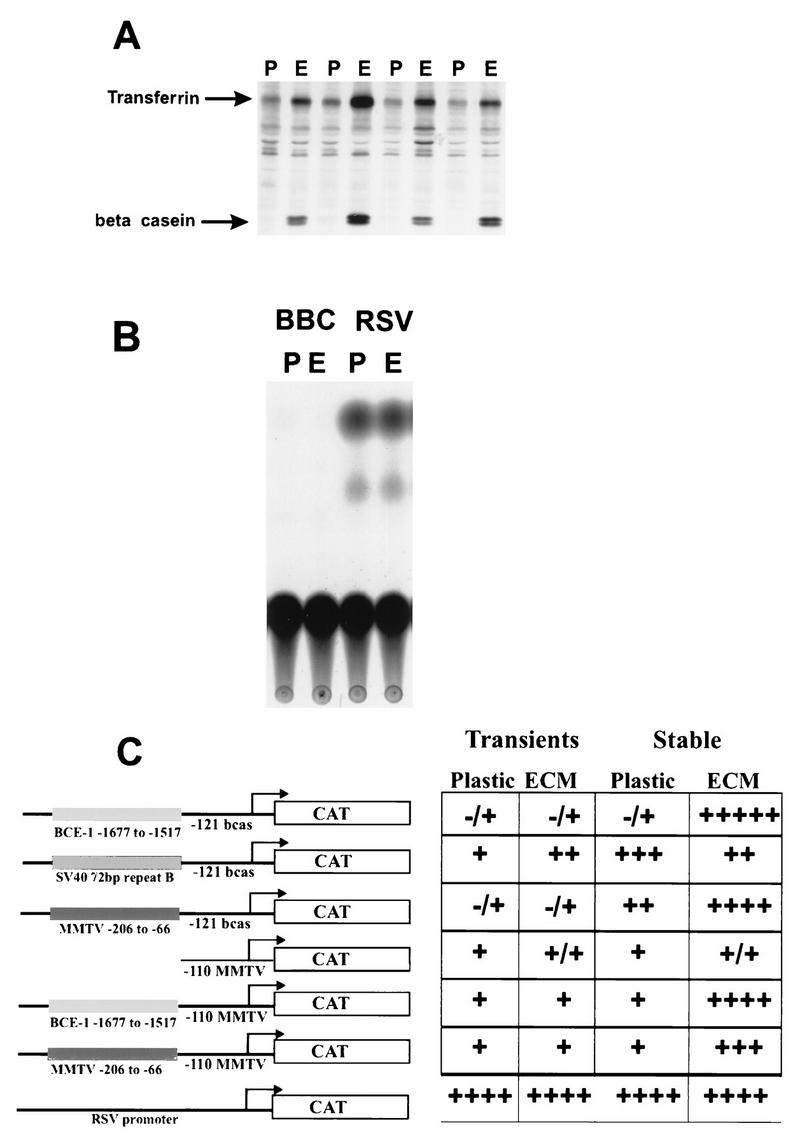
ECM does not induce transcriptional activation of nonintegrated templates in transient-transfection analysis. (A) Autoradiograph of an immunoprecipitation reaction using an antibody to mouse milk. The 35S-labeled proteins were precipitated from parallel plates of transiently transfected cells cultured on plastic or low-density polyhema treated with 2% ECM. (B) A representative autoradiograph of a CAT assay. Ten micrograms of protein from cell lysates isolated from CID-9 cells transiently transfected with BBC or RSV/CAT while cultured on plastic (P) or low-density polyhema treated with 2% ECM (E) was analyzed. (C) A diagram for each plasmid tested in the transient-transfection analysis is shown to the left. BCE-1, β-casein, and MMTV are labeled relative to the transcription start site of the endogenous genes. The seven constructs contained the reporter CAT gene. The adjacent table represents the activity of each construct when transiently or stably transfected into CID-9 cells and differentiated in IHP on plastic or ECM as described in Materials and Methods. The transient activity is relative to cotransfected RSV–β-Gal expression (a promoter which is not regulated by the presence of ECM). −, no detectable activity; +, increasing amounts of transcriptional activity. These data represent at least three independent transfections for each condition.
Inhibitors of histone deacetylase induce BCE-1-mediated transcription in the absence of ECM.
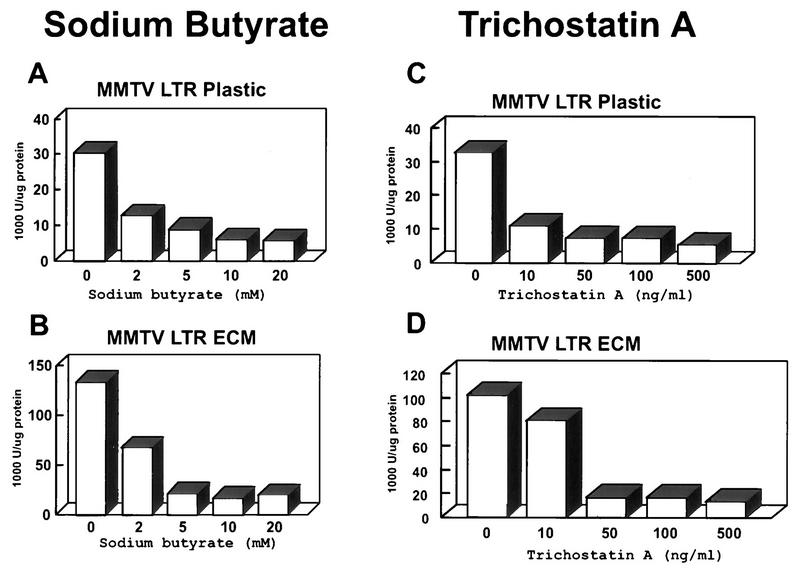
Stably transfected CID-9 cells cultured on tissue culture plastic or ECM were treated with sodium butyrate, an inhibitor of histone deacetylase. Treatment with sodium butyrate is capable of inducing transcription to levels achieved in the presence of ECM (Fig. 6A). The induction of transcription by sodium butyrate was reproducible with differences in the absolute fold increase in the absence of ECM. (Figure 6B shows a representative CAT TLC and a graph of the quantitated results.) However, in the presence of ECM, when transcription is high, the transcriptional induction by sodium butyrate was always equal to or less than twofold (Fig. 6A and data not shown). Since sodium butyrate treatment has been shown to induce many changes in the cell in addition to the inhibition of histone deacetylase (18, 24, 27, 40), we tested a more specific inhibitor of histone deacetylase, trichostatin A (62). A dose-dependent induction of transcription of up to 10-fold was observed in cells cultured with trichostatin A in the absence of ECM (Fig. 6C) which, as with sodium butyrate, achieved levels similar to that of the ECM induction and induced ECM levels less than twofold (data not shown). To determine whether sodium butyrate and trichostatin A activation was due to a generalized increase in cellular transcription, we analyzed the ECM-responsive MMTV promoter in stably transfected CID-9 cells under the same culture conditions and treatments. The presence of sodium butyrate, as well as of trichostatin A, led to a dose-dependent repression of MMTV transcription either in the absence (Fig. 7A and C) or in the presence (Fig. 7B and D) of ECM. The activation of BCE-1 and the inactivation of MMTV through inhibition of histone deacetylase suggest that ECM-dependent transcription may involve alterations in the acetylation state of the histones.
FIG. 6.
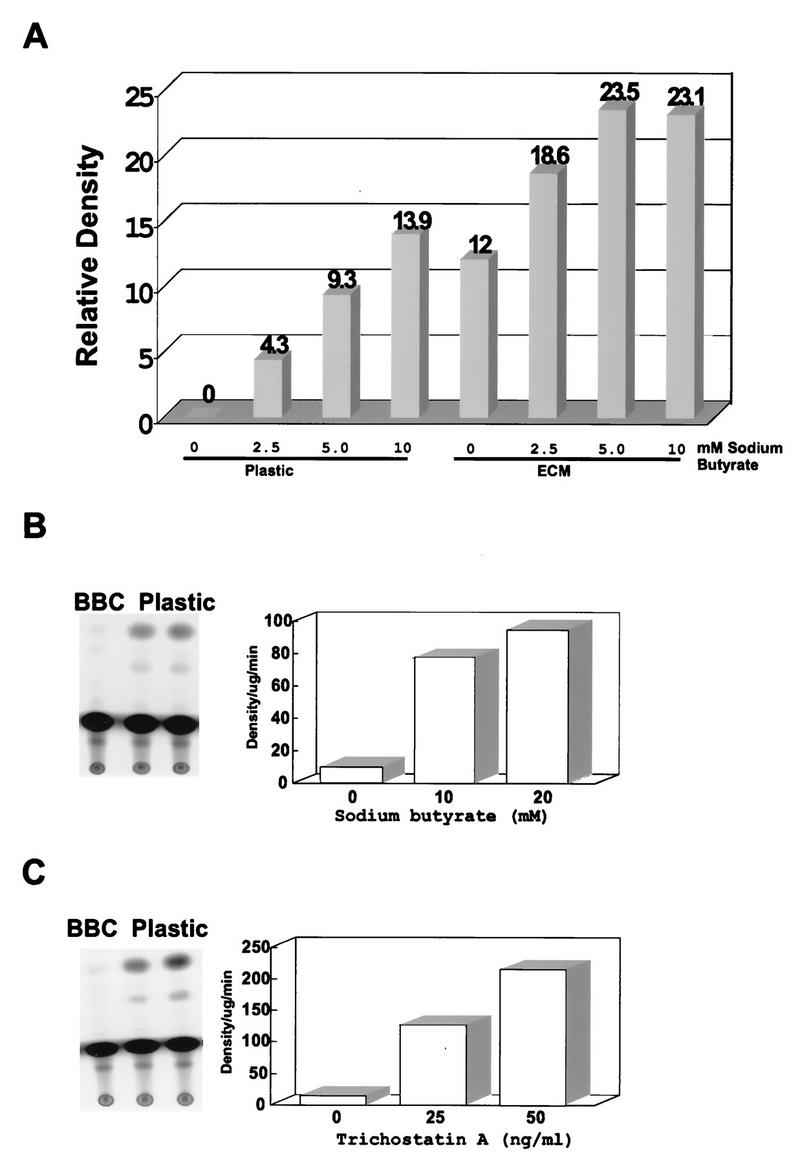
Treatment of CID-9 cells with histone deacetylase inhibitors induces transcription in the absence of ECM in stable BBC transfectants. (A) CID-9 cells were cultured on plastic or ECM and treated with various doses of sodium butyrate. Cell lysates were assayed for CAT activity, and the graph represents the densities relative to those of the cells on plastic with no treatment. (B and C) Representatives of at least three independent experiments where stably BBC-transfected CID-9 cells were plated on plastic in the presence of differentiation medium for 2 days before harvest. Sodium butyrate or trichostatin A was added 18 h before harvest. The autoradiograph shows a TLC separation of the CAT reaction products, utilizing 10 μg of protein from cell lysates for the assay. The graph represents density per microgram of protein per minute of CAT reaction. (The treatments described above will induce BBC activity in cells cultured on ECM by about twofold; data not shown.)
FIG. 7.
Treatment of CID-9 cells with histone deacetylase inhibitors has opposing effects on the stable MMTV transfectants. Representative results from one of at least three experiments where stable MMTV LTR transfectants were plated on plastic or ECM in differentiation medium. The cells were cultured and treated with sodium butyrate (A and B) or with trichostatin A (C and D) as described for Fig. 6. The data is expressed as units of luciferase activity per microgram of protein.
DISCUSSION
In this paper we show that transcriptional activation of β-casein, via the BCE-1 enhancer, requires that the ECM and prolactin coordinate the interactions between at least three transcription factors and the chromosomally integrated template containing BCE-1. Furthermore, BCE-1 transcriptional activation can be induced in the absence of ECM upon treatment of stably transfected CID-9 cells with inhibitors of histone deacetylase.
Mutation of any of three regions within BCE-1 abolishes ECM- and prolactin-induced transcription, suggesting a cooperative interplay between the factors that bind to these sites. Our analyses indicate that C/EBPβ, STAT5, and an additional but as-yet-unknown protein (i.e., the UBP) recognize these sites. Several laboratories have demonstrated the requirement for both STAT and C/EBP for hormone-induced β-casein expression. Standke et al. (49) have shown that truncation of the rat β-casein promoter, which removes C/EBP binding sites and leaves only the STAT site, abolishes induction of β-casein transcription. Doppler et al. (16) have shown by truncation analysis that both the C/EBP and STAT sites of the rat β-casein promoter are required for activation of a heterologous thymidine kinase promoter. Taken together, these experiments suggest that the induction of β-casein expression minimally requires the interplay of C/EBP and STAT and, in the case of BCE-1, the UBP.
The ECM, in which laminin has been shown to be the active component, induces β-casein gene expression requiring each of the following: a rounded cell shape (42), signal transduction via laminin binding and ligating of a β1-containing integrin (52), prolactin, integration into the genome, and a promoter which contains intact binding sites for C/EBP, STAT5, and the UBP. Furthermore, ECM-induced β-casein expression may require a modulation of the state of histone acetylation. However, the ECM does not appear to modulate the levels or binding activities of nuclear proteins specific to the C/EBP, STAT5, and UBP binding sites. In vitro EMSA and in vivo exonuclease footprinting analysis (in the case of C/EBP) indicate that these factors are present and able to bind DNA in the absence of ECM. This is in contrast to the ECM-dependent induction of albumin expression in hepatocytes, where liver-specific gene transcription appears to depend on the presence of liver-enriched transcription factors which are upregulated in hepatocytes cultured on ECM (13).
The findings that STAT5 has binding activity (Fig. 3) and that a STAT5 reporter gene construct is activated to the same extent (data not shown) whether CID9 cells were cultured under differentiation conditions or not differ from the results obtained by Streuli et al. (51). Streuli et al. showed that nuclear extracts from primary mouse mammary epithelial cells cultured on ECM formed a specific complex with a STAT5 binding site in the β-lactoglobulin promoter, whereas extracts from cells cultured on plastic did not. This binding activity was maintained only in cells cultured on ECM in the presence of prolactin. While it is possible that the binding of STAT5 differs for sheep β-lactoglobulin and bovine β-casein, another explanation for this discrepancy could reside in differences between primary and immortalized cells. The primary cells undoubtedly have complex requirements for induction of expression of tissue-specific genes which may be relaxed or altered in cell lines. Indeed the homolog, STAT5b, is able to activate transcription of the rat β-casein gene in COS cells cultured in the absence of prolactin (32). Constitutively activated STAT5 has been observed in T cells transformed with human T-cell leukemia virus type 1 (35) and in peripheral blood cells from patients with acute leukemia (21). In addition, STAT binding to the interleukin-6 response element and the hematopoietin receptor response element has been observed in the absence of transcriptional activation (26). It is therefore plausible that in immortalized CID-9 cells STAT5 could be present and constitutively bound to BCE-1 in the absence of ECM. These cell lines now make it possible to study the events subsequent to binding of STAT, which would not be possible in primary cultures.
Thus, the experiments we report here lead us to conclude that the binding of STAT5 is not sufficient to activate transcription of the BCE-1 enhancer in the absence of ECM and prolactin. The fact that the other factors also appear to be bound in the absence of ECM indicates that the transcriptional activation is not solely due to the binding of individual transcription factors. Rather, a proper structural signal may be required for the activation of preassembled transcriptional machinery which is poised for rapid transcription. This observation has a precedent in the literature: in vivo footprinting analysis reveals that the occupancy of the sites in the serum response element of the fos promoter remains unchanged regardless of the transcriptional state of the gene (22). In that study the authors proposed and subsequently showed (64) that activation and inactivation may be brought about by modifications to the proteins in the bound complex or by interactions between the complex and additional factors. The continuous occupancy of the serum response element may be a mechanism for achieving a rapid activation as maximal fos transcription occurs less than 20 min after growth factor addition. Although β-casein is not considered a rapid-response gene, it is conceivable that committed mammary epithelial cells have been organized in vivo to respond rapidly to conditions that modulate milk protein gene expression. Therefore, it would be interesting to determine the occupancy of factors bound to BCE-1 in nonmammary epithelial cells.
In vitro factor binding analyses indicate that the factors which are critical for transcriptional activation via BCE-1 are present and are capable of binding in both the transcriptionally active and inactive states. Although factor binding does not address the functionality of proteins bound to the DNA or whether the composition of the bound complex has changed, the lack of response to ECM by nonintegrated BCE-1 constructs suggests that modifications of trans-acting factors by ECM or changes in the complex composition are not sufficient to account for the enhancer’s function and that modifications within the context of chromatin are necessary. Cell type-specific chromatin arrangements mediated via cell-specific factors have been described for the liver-specific albumin gene in hepatocytes (34). The albumin enhancer exists in an array of three positioned nucleosomes only in hepatocytes, and this positioning is dependent upon binding of a liver-enriched nuclear factor, HNF3.
Chromatin alterations which lead to induction of gene expression may be achieved through any or all of the following: modifications of histones, changes in DNA conformation, factor accessibility, and DNA and/or nuclear factor localization (for general reviews, see references 17 and 23). The fact that integrated BCE-1 is activated in the absence of ECM upon treatment with inhibitors of histone deacetylase suggests that a component of ECM-dependent β-casein expression involves histone modifications, which are believed to be involved in the transcriptional regulation of genes (reviewed in references 33, 55, and 60). One possible mechanism involves ECM-induced changes in the three-dimensional architecture of the cell, influencing in turn the three-dimensional architecture of the nucleus. Alterations in the structure or composition of the nuclear matrix may reposition histone acetyltransferases and/or deacetylases, which are known to be bound to the nuclear scaffolding (9). The changes in protein-nuclear scaffold and/or protein-DNA interactions involved in this nuclear reorganization may result in the activation of the β-casein gene following a more direct change in the modification state of the histones (see reference 4 and references therein for hypothesis). Another possibility is that the ECM induces or modifies cofactors which themselves have acetyltransferase or deacetylase activity. Recent characterization of novel histone acetyltransferases includes p300/CBP-associated factor (61). The transcriptional coactivator p300 (38) and TAFII250 (37) suggest potential targets which could be involved in ECM-induced transcription. Interestingly, p300 directly interacts with C/EBPβ to increase transcriptional activity (36) and also interacts with the transcriptional repressor YY1 to relieve YY1 repression (28). These findings are consistent with the current understanding of mechanisms underlying the transcriptional regulation of β-casein.
Although factor availability and modification of DNA accessibility of C/EBP do not account for ECM-dependent activation of the BCE-1 element, it is clear that complex interactions between several factors on a chromosomally integrated template are involved. Whether the ECM-induced changes in higher-order chromatin structures are mediated via alterations in nuclear architecture and/or ECM induces cofactors which modulate the modification of the state of histones is currently under investigation.
ACKNOWLEDGMENTS
We are grateful to Steve Farmer for providing the protocol for ECM dissolution, Lothar Henninghausen for the STAT5a STAT5b antibodies, and Wolfgang Doppler for helpful scientific discussions. We thank Paul Kaufman, Judy Campisi, Nancy Boudreau, and Niveen Malek for critically reading the manuscript.
C. Schmidhauser and R. Mossi were partly supported by the Swiss Cancer League. A majority of this work was supported by the office of Health and Environmental Research of the U.S. Department of Energy (under contract number DE-AC03-SF00098 to M.J.B.). This work was also supported by a gift from Monsanto Company and a grant from the U.S.-Israel Binational Agricultural Research and Development Fund (BARD project no. IS-2373-94R) to M.J.B.
REFERENCES
- 1.Adams J C, Watt F M. Regulation of development and differentiation by the extracellular matrix. Development. 1993;117:1183–1198. doi: 10.1242/dev.117.4.1183. [DOI] [PubMed] [Google Scholar]
- 2.Barcellos-Hoff M H, Aggeler J, Ram T G, Bissell M J. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development. 1989;105:223–235. doi: 10.1242/dev.105.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bissell M J, Hall H G. The role of extracellular matrix. In: Nevelle M, Daniel C, editors. The mammary gland. New York, N.Y: Plenum Press; 1987. pp. 97–146. [Google Scholar]
- 4.Boudreau N J, Myers C A, Bissell M J. From laminin to lamin: regulation of tissue-specific gene expression by the ECM. Trends Cell Biol. 1995;5:1–4. doi: 10.1016/s0962-8924(00)88924-2. [DOI] [PubMed] [Google Scholar]
- 5.Burdon T G, Demmer J, Clark A J, Watson C J. The mammary factor MPBF is a prolactin-induced transcriptional regulator which binds to STAT factor recognition sites. FEBS Lett. 1994;350:177–182. doi: 10.1016/0014-5793(94)00757-8. [DOI] [PubMed] [Google Scholar]
- 6.Burdon T G, Maitland K A, Clark A J, Wallace R, Watson C J. Regulation of the sheep beta-lactoglobulin gene by lactogenic hormones is mediated by a transcription factor that binds an interferon-gamma activation site-related element. Mol Endocrinol. 1994;8:1528–1536. doi: 10.1210/mend.8.11.7877621. [DOI] [PubMed] [Google Scholar]
- 7.Cao Z, Umek R M, McKnight S L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 8.Danielson K G, Oborn C J, Durban E M, Buetel J S, Medina D. Epithelial mouse mammary cell line exhibiting normal morphogenesis in vivo and functional differentiation in vitro. Proc Natl Acad Sci USA. 1984;81:3756–3760. doi: 10.1073/pnas.81.12.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davie J R. Histone modifications, chromatin structure, and the nuclear matrix. J Cell Biochem. 1996;62:149–157. doi: 10.1002/(sici)1097-4644(199608)62:2<149::aid-jcb2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 10.Deryckere F, Gannon F. A one-hour minipreparation technique for extraction of DNA-binding proteins from animal tissues. BioTechniques. 1994;16:405. [PubMed] [Google Scholar]
- 11.Desprez P, Roskelley C D, Campisi J, Bissell M J. Isolation of functional cell lines from a mouse mammary epithelial cell strain: the importance of basement membrane and cell-cell interaction. Mol Cell Differ. 1993;1:99–110. [Google Scholar]
- 12.de Wet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiPersio C M, Jackson D A, Zaret K S. The extracellular matrix coordinately modulates liver transcription factors and hepatocyte morphology. Mol Cell Biol. 1991;11:4405–4414. doi: 10.1128/mcb.11.9.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doppler W, Groner B, Ball R K. Prolactin and glucocorticoid hormones synergistically induce expression of transfected rat beta-casein gene promoter constructs in a mammary epithelial cell line. Proc Natl Acad Sci USA. 1989;86:104–108. doi: 10.1073/pnas.86.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doppler W, Hock W, Hofer P, Groner B, Ball R K. Prolactin and glucocorticoid hormones control transcription of the beta-casein gene by kinetically distinct mechanisms. Mol Endocrinol. 1990;4:912–919. doi: 10.1210/mend-4-6-912. [DOI] [PubMed] [Google Scholar]
- 16.Doppler W, Welte T, Philipp S. CCAAT/enhancer-binding protein isoforms beta and delta are expressed in mammary epithelial cells and bind to multiple sites in the beta-casein gene promoter. J Biol Chem. 1995;270:17962–17969. doi: 10.1074/jbc.270.30.17962. [DOI] [PubMed] [Google Scholar]
- 17.Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 18.Ginsburg E, Salomon D, Sreevalsan T, Freese E. Growth inhibition and morphological changes caused by lipophilic acids in mammalian cells. Proc Natl Acad Sci USA. 1973;70:2457–2461. doi: 10.1073/pnas.70.8.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman H S, Rosen J M. Transcriptional analysis of the mouse beta-casein gene. Mol Endocrinol. 1990;4:1661–1670. doi: 10.1210/mend-4-11-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouilleux F, Wakao H, Mundt M, Groner B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J. 1994;13:4361–4369. doi: 10.1002/j.1460-2075.1994.tb06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gouilleux-Gruart V, Gouilleux F, Desaint C, Claisse J F, Capiod J C, Delobel J, Weber-Nordt R, Dusanter-Fourt I, Dreyfus F, Groner B, Prin L. STAT-related transcription factors are constitutively activated in peripheral blood cells from acute leukemia patients. Blood. 1996;87:1692–1697. [PubMed] [Google Scholar]
- 22.Herrera R E, Shaw P E, Nordheim A. Occupation of the c-fos serum response element in vivo by a multi-protein complex is unaltered by growth factor induction. Nature. 1989;340:68–70. doi: 10.1038/340068a0. [DOI] [PubMed] [Google Scholar]
- 23.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 24.Kruh J. Effects of sodium butyrate, a new pharmacological agent, on cells in culture. Mol Cell Biochem. 1982;42:65–82. doi: 10.1007/BF00222695. [DOI] [PubMed] [Google Scholar]
- 25.Kulski J K, Topper Y J, Chomczynski P, Qasba P. An essential role for glucocorticoid in casein gene expression in rat mammary explants. Biochem Biophys Res Commun. 1983;114:380–387. doi: 10.1016/0006-291x(83)91638-8. [DOI] [PubMed] [Google Scholar]
- 26.Lai C F, Ripperger J, Morella K K, Wang Y, Gearing D P, Fey G H, Bauman H. Separate signaling mechanisms are involved in the control of STAT protein activation and gene regulation via interlurkin 6 response element by the box 3 motif of gp130. J Biol Chem. 1995;270:14847–14850. doi: 10.1074/jbc.270.25.14847. [DOI] [PubMed] [Google Scholar]
- 27.Leder A, Leder P. Butyric acid, a potent inducer of erythroid differentiation in cultured erythroleukemic cells. Cell. 1975;5:319–322. doi: 10.1016/0092-8674(75)90107-5. [DOI] [PubMed] [Google Scholar]
- 28.Lee J S, Galvin K M, See R H, Eckner R, Livingston D, Moran E, Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 29.Lee K F, DeMayo F J, Atiee S H, Rosen J M. Tissue-specific expression of the rat beta-casein gene in transgenic mice. Nucleic Acids Res. 1988;16:1027–1041. doi: 10.1093/nar/16.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee P P, Lee M T, Darcy K M, Shudo K, Ip M M. Modulation of normal mammary epithelial cell proliferation, morphogenesis, and functional differentiation by retinoids: a comparison of the retinobenzoic acid derivative RE80 with retinoic acid. Endocrinology. 1995;136:1707–1717. doi: 10.1210/endo.136.4.7895682. [DOI] [PubMed] [Google Scholar]
- 31.LeFebvre P, Berard D S, Cordingley M G, Hager G L. Two regions of the mouse mammary tumor virus long terminal repeat regulate the activity of its promoter in mammary cell lines. Mol Cell Biol. 1991;11:2529–2537. doi: 10.1128/mcb.11.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Robinson G, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc Natl Acad Sci USA. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loidl P. Histone acetylation: facts and questions. Chromosoma. 1994;103:441–449. doi: 10.1007/BF00337382. [DOI] [PubMed] [Google Scholar]
- 34.McPherson C E, Shim E Y, Friedman D S, Zaret K S. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell. 1993;75:387–398. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- 34a.Michelotti, J. Unpublished data.
- 35.Migone T S, Lin J X, Cereseto A, Mulloy J C, O’Shea J J, Franchini G, Leonard W J. Constitutively activated Jak-STAT pathway in T cells transformed with HTLV-I. Science. 1995;269:79–81. doi: 10.1126/science.7604283. [DOI] [PubMed] [Google Scholar]
- 36.Mink S, Haenig B, Klempnauer K H, Spemann H. Interaction and functional collaboration of p300 and C/EBPβ. Mol Cell Biol. 1997;11:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizzen C A, Yang X, Kokubo T, Brownell J E, Bannister A J, Owen-Huges T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 37a.Myers, C., and J. Michelotti. Unpublished data.
- 38.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;5:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 39.Pennie W D, Hager G L, Smith C L. Nucleoprotein structure influences the response of the mouse mammary tumor virus promoter to activation of the cyclic AMP signalling pathway. Mol Cell Biol. 1995;15:2125–2134. doi: 10.1128/mcb.15.4.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prasad K N, Sinha P K. Effect of sodium butyrate on mammalian cells in culture: a review. In Vitro. 1976;12:125–132. doi: 10.1007/BF02796360. [DOI] [PubMed] [Google Scholar]
- 41.Raught B, Liao W S, Rosen J M. Developmentally and hormonally regulated CCAAT/enhancer-binding protein isoforms influence beta-casein gene expression. Mol Endocrinol. 1995;9:1223–1232. doi: 10.1210/mend.9.9.7491114. [DOI] [PubMed] [Google Scholar]
- 42.Roskelley C D, Desprez P Y, Bissell M J. Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc Natl Acad Sci USA. 1994;91:12378–12382. doi: 10.1073/pnas.91.26.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roskelley C D, Srebrow A, Bissell M J. A hierarchy of ECM-mediated signalling regulates tissue-specific gene expression. Curr Opin Cell Biol. 1995;7:736–747. doi: 10.1016/0955-0674(95)80117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidhauser C, Bissell M J, Myers C A, Casperson G F. Extracellular matrix and hormones transcriptionally regulate bovine beta-casein 5′ sequences in stably transfected mouse mammary cells. Proc Natl Acad Sci USA. 1990;87:9118–9122. doi: 10.1073/pnas.87.23.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidhauser C, Casperson G F, Bissell M J. Transcriptional activation by viral enhancers: critical dependence on extracellular matrix-cell interactions in mammary epithelial cells. Mol Carcinog. 1994;10:66–71. doi: 10.1002/mc.2940100203. [DOI] [PubMed] [Google Scholar]
- 46.Schmidhauser C, Casperson G F, Myers C A, Sanzo K T, Bolten S, Bissell M J. A novel transcriptional enhancer is involved in the prolactin- and extracellular matrix-dependent regulation of beta-casein gene expression. Mol Biol Cell. 1992;3:699–709. doi: 10.1091/mbc.3.6.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitt-Ney M, Doppler W, Ball R K, Groner B. Beta-casein gene promoter activity is regulated by the hormone-mediated relief of transcriptional repression and a mammary-gland-specific nuclear factor. Mol Cell Biol. 1991;11:3745–3755. doi: 10.1128/mcb.11.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shapiro D J, Sharp P A, Wahli W W, Keller M J. A high-efficiency HeLa cell nuclear transcription extract. DNA. 1988;7:47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- 49.Standke G J, Meier V S, Groner B. Mammary gland factor activated by prolactin on mammary epithelial cells and acute-phase response factor activated by interleukin-6 in liver cells share DNA binding and transactivation potential. Mol Endocrinol. 1994;8:469–477. doi: 10.1210/mend.8.4.7519723. [DOI] [PubMed] [Google Scholar]
- 50.Stocklin E, Wissler M, Gouilleux F, Groner B. Functional interactions between Stat 5 and the glucocorticoid receptor. Nature. 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- 51.Streuli C H, Edwards G M, Delcommenne M, Whitelaw C B, Burdon T G, Schindler C, Watson C J. Stat5 as a target for regulation by extracellular matrix. J Biol Chem. 1995;270:21639–21644. doi: 10.1074/jbc.270.37.21639. [DOI] [PubMed] [Google Scholar]
- 52.Streuli C H, Schmidhauser C, Bailey N, Yurchenco P, Skubitz A P, Roskelley C, Bissell M J. Laminin mediates tissue-specific gene expression in mammary epithelia. J Cell Biol. 1995;129:591–603. doi: 10.1083/jcb.129.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taketani Y, Oka T. Tumor promoter 12-O-tetradecanoylphorbol 13-acetate, like epidermal growth factor, stimulates cell proliferation and inhibits differentiation of mouse mammary epithelial cells in culture. Proc Natl Acad Sci USA. 1983;80:1646–1649. doi: 10.1073/pnas.80.6.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Topper Y J, Freeman C S. Multiple hormone interactions in the developmental biology of the mammary gland. Physiol Rev. 1980;60:1049–1106. doi: 10.1152/physrev.1980.60.4.1049. [DOI] [PubMed] [Google Scholar]
- 55.Turner B M, O’Neill L P. Histone acetylation in chromatin and chromosomes. Semin Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- 56.Wakao H, Gouilleux F, Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J. 1994;13:2182–2191. doi: 10.1002/j.1460-2075.1994.tb06495.x. . (Erratum, 14:854–855, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wakao H, Gouilleux F, Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J. 1995;14:854–855. doi: 10.1002/j.1460-2075.1995.tb07064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watson C J, Gordon K E, Robertson M, Clark A J. Interaction of DNA-binding proteins with a milk protein gene promoter in vitro: identification of a mammary gland-specific factor. Nucleic Acids Res. 1991;19:6603–6610. doi: 10.1093/nar/19.23.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Welte T, Garimorth K, Philipp S, Doppler W. Prolactin-dependent activation of a tyrosine phosphorylated DNA binding factor in mouse mammary epithelial cells. Mol Endocrinol. 1994;8:1091–1102. doi: 10.1210/mend.8.8.7527899. [DOI] [PubMed] [Google Scholar]
- 60.Wolffe A P. Histone deacetylase: a regulator of transcription. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 61.Yang X J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 62.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 63.Yoshimura M, Oka T. Transfection of beta-casein chimeric gene and hormonal induction of its expression in primary murine mammary epithelial cells. Proc Natl Acad Sci USA. 1990;87:3670–3674. doi: 10.1073/pnas.87.10.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zinck R, Hipskind R A, Pingoud V, Nordheim A. c-fos transcriptional activation and repression correlate temporally with the phosphorylation status of TCF. EMBO J. 1993;12:2377–2387. doi: 10.1002/j.1460-2075.1993.tb05892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]