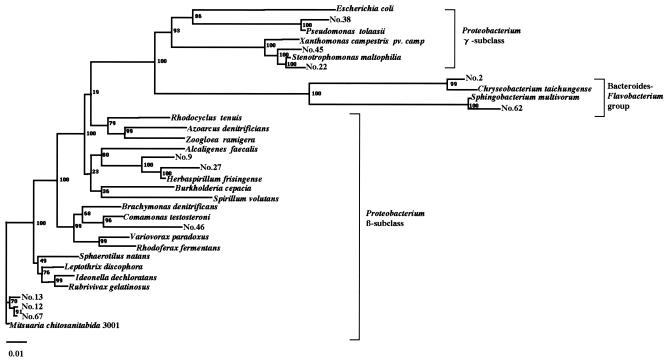
FIG. 3.
Phylogenetic relationship of M. chitosanitabida 3001 with the 11 selected isolates and their related strains based on their 16S rRNA gene sequences. Bar, 1 nucleotide substitution per 100 nucleotides in the 16S rRNA gene sequence. The numbers at the nodes of the tree indicate bootstrap values (percentages) for each node of 1,000 bootstrap resamplings. The sequences used for the comparison with the 16S rRNA genes of the isolates were obtained from GenBank. The origins and accession numbers of the sequences are as follows: Escherichia coli, J01859; Pseudomonas sp. strain 38, AY856844; Pseudomonas tolaasii, AF255336; Xanthomonas campestris pv. campestris, AF000946; Stenotrophomonas sp. strain 45, AY856845; Stenotrophomonas maltophilia, AJ131903; Stenotrophomonas sp. strain 22, AY856842; Chryseobacterium sp. strain 2, AB024308; Chryseobacterium taichungense, AJ843132; Sphingobacterium multivorum, AB020205; Sphingobacterium sp. strain 62, AY856847; Rhodocyclus tenuis, D16208; Azoarcus denitrificans, L33694; Zoogloea ramigera, D14257; Alcaligenes faecalis, D88008; Herbaspirillum sp. strain 9, AB024305; Herbaspirillum sp. strain 27, AY856843; Herbaspirillum frisingense, AJ238359; Burkholderia cepacia, X87275; Spirillum volutans, M34131; Brachymonas denitrificans, D14320; Comamonas testosteroni, AB064318; Comamonas sp. strain 46, AY856846; Variovorax paradoxus, D88006; Rhodoferax fermentans, D16212; Sphaerotilus natans, Z18534; Leptothrix discophora, Z18533; Ideonella dechloratans, X72724; Rubrivivax gelatinosus, AB016167; Mitsuaria sp. strain 13, AB024306; Mitsuaria chitosanitabida 3001, AB024307; Mitsuaria sp. strain 67, AY856848; Mitsuaria sp. strain 12, AY856841.