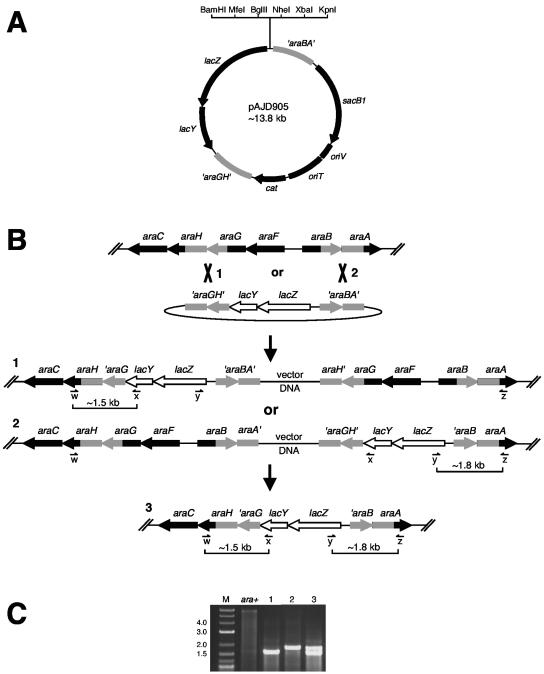
FIG. 1.
Integration of lacZY operon fusions into the ara locus. (A) Partial restriction map of plasmid pAJD905. Restriction sites shown in the polylinker are unique (note that MfeI is compatible with EcoRI). (B) Summary of lacZY fusion integration procedure (see text for experimental details). Plasmid pAJD905 is integrated into the ara locus by homologous recombination between either the ′araGH′ sequences or the ′araBA′ sequences. The structures of both possible integrants are shown as structures 1 and 2. Next, sucrose-resistant segregants lose the integrated pAJD905 backbone by homologous recombination between the tandemly duplicated ara sequences. This either regenerates the ara+ locus or leaves the lacZY fusion on the chromosome, shown as structure 3. (C) Colony PCR analysis of the ara+ parental strain, integrants with structure 1 or 2, and segregant 3 is shown. In this case the lacZY operon from pAJD905 without a promoter cloned upstream was used. Binding sites of each primer are shown in panel B as w, x, y, and z (see also Table 2). For the ara+ strain, primers w and z generate a single product of ∼6 kb that is only weakly amplified (primers x and y do not anneal). Integrants 1 and 2 give only one PCR product, as indicated in panel B (the second pair of primers are too far apart for amplification). The desired segregant gives two PCR products as shown. Note that a promoter fragment inserted upstream of the lacZY operon would increase the size of the 1.8-kb product. M, DNA marker (kilobase sizes of some marker bands are indicated). The gel lane images have been rearranged from the original order, but the data are from the same gel and a single experiment.