FIG. 1.
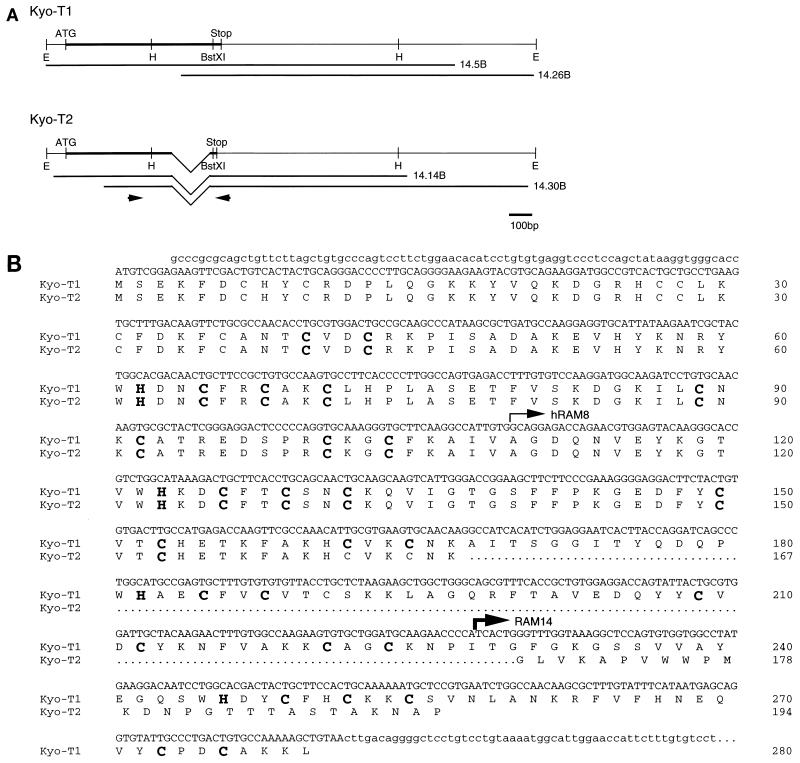
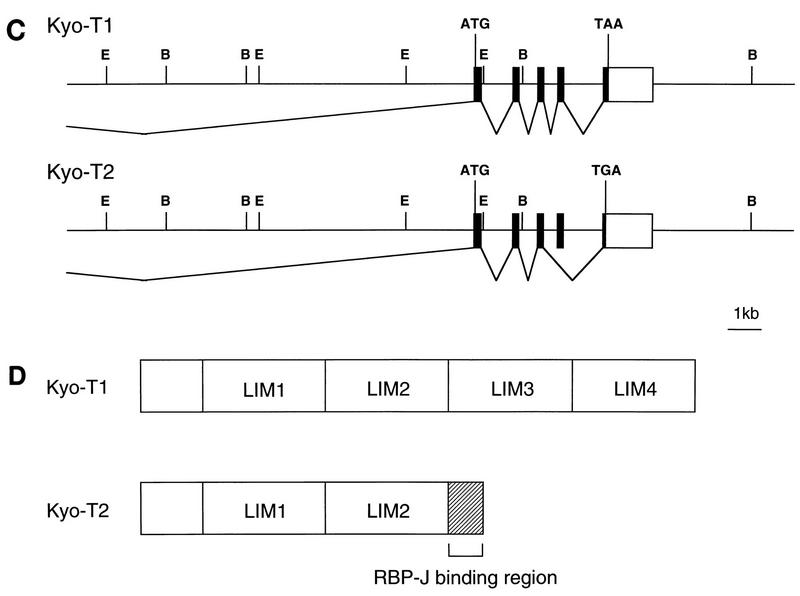
Sequence and structure of KyoT. (A) cDNA structure of KyoT. The coding regions are represented by thick lines. The cDNA fragments obtained from the mouse brain cDNA library are shown below the putative full-length cDNAs. The positions of primers used for RT-PCR are shown by arrows. E, EcoRI; H, HindIII; ATG, putative initiation codon; Stop, termination codon. (B) Nucleotide and deduced amino acid sequences of KyoT. Nucleotide sequences of coding and noncoding regions are shown as upper- and lowercase letters, respectively. The first 167 amino acids are shared between two alternatively spliced isoforms, KyoT1 and KyoT2. The amino acids removed by alternative splicing in KyoT2 are represented by dots. Four LIM domains are boxed, and the conserved cysteines and histidines within the LIM motif are shown as boldface letters. The 5′ ends of the fragments obtained by two-hybrid screening from mouse embryo (RAM14) and HeLa (hRAM8) cDNA libraries are shown by thin and thick arrows, respectively. (C) Genomic structure of KyoT. The functional gene is composed of at least five exons and spans at least 20 kbp. In KyoT2 mRNA, one of the exons is removed by alternative splicing. Most of the 5′ untranslated region was not cloned. Solid boxes, coding region; open boxes, untranslated region; ATG, first methionine of the coding region; TAA and TAG, termination codons of KyoT1 and KyoT2, respectively; E, EcoRI; B, BamHI. (D) Simplified diagram showing the structures of the KyoT1 and KyoT2 proteins. The LIM domains are numbered from the N terminus.