Summary
Histological analysis of intestinal epithelial tissues is enhanced by 3D visualization compared to 2D sections. Here, we present a protocol for 3D visualization of intestinal epithelial cells using an optical clearing approach optimized for endogenous fluorescence and preservation of crypt-villus morphology. We describe steps for clearing and refractive index matching tissue. We provide detailed procedures for imaging and reconstructing tissue to visualize epithelial cells along the crypt-villus axis with high resolution. We illustrate this approach with endogenous tdTomato used for lineage tracing in the small intestine of Fgfbp1-CreERT2; Rosa26-tdTomato mice.
For complete details on the use and execution of this protocol, please refer to Capdevila et al.1
Subject area: Cell Biology, Microscopy, Stem Cells, Cell Differentiation
Graphical abstract

Highlights
-
•
Instructions for optical clearing and 3D reconstruction of murine small intestine
-
•
Steps to delipidate tissues to visualize lineage traces without losing structural integrity
-
•
Guidance on 3D visualization without using a light sheet microscope
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Histological analysis of intestinal epithelial tissues is enhanced by 3D visualization compared to 2D sections. Here, we present a protocol for 3D visualization of intestinal epithelial cells using an optical clearing approach optimized for endogenous fluorescence and preservation of crypt-villus morphology. We describe steps for clearing and refractive index matching tissue. We provide detailed procedures for imaging and reconstructing tissue to visualize epithelial cells along the crypt-villus axis with high resolution. We illustrate this approach with endogenous tdTomato used for lineage tracing in the small intestine of Fgfbp1-CreERT2; Rosa26-tdTomato mice.
Before you begin
Lgr5+ crypt base columnar (CBC) cells are widely considered to be the intestinal stem cells (ISCs) responsible for homeostatic epithelial regeneration, producing progeny that migrate upward unidirectionally through the poorly understood transit-amplifying (TA) zone in the upper crypt before differentiating into villus cells.2 However, targeted deletion of Lgr5+ cells using diphtheria toxin (DT) or pharmacological suppression of Lgr5+ cells by R-spondin signaling blockade shows that crypt proliferation and epithelial homeostasis persist despite Lgr5+ CBC cell depletion, indicating the presence of other regenerative populations.3,4 Our lab recently identified a novel ISC population of Fgfbp1+ cells located in the upper crypt, distinct from Lgr5+ cells. These cells contribute to epithelial regeneration in a bi-directional manner, generating mature lineages in the villus while also replenishing Lgr5+ cells at the crypt base.1
To characterize Fgfbp1+ cells and their progeny, we employed an Fgfbp1-CreERT2 knock-in allele for tamoxifen (TAM)-induced lineage tracing in Fgfbp1-CreERT2; Rosa26-tdTomato mice, revealing tdTomato+ labeling of upper crypt cells within 18 h of TAM induction.1 Given the necessity for precise visualization, we utilized an optical clearing protocol to enable 3D imaging of the intestinal epithelium. This approach allowed for the accurate tracking of Fgfbp1+ cell kinetics, spatial distribution, and lineage contributions confirming their role as ISCs. By integrating optical clearing, fluorescent labeling, and 3D reconstruction, we provided a newfound perspective on the dynamics of these stem cells within the intestinal upper crypt.
Among tissue clearing techniques, CLARITY was developed to enable optical transparency by removing lipids while preserving key biomolecules and tissue structures.5 This allows for deep tissue imaging with fluorescent labeling and immunostaining, making it valuable for high-resolution 3D analysis.5 Recent advancements in optical clearing techniques have significantly improved the visualization of intestinal tissues. Various methods have been successfully applied to clear and image mouse intestines, including the aqueous clearing agent FocusClear,6,7,8 urea-based ScaleA2,8 fructose-based SeeDB,9 Click-It Chemistry for fluorescence detection,10 CUBIC,11 Murray’s Clear,12 and Clearing-enhanced 3D (Ce3D).13,14 More recently, the PACT variation of the CLARITY method has been utilized to study the enteric nervous system in both human and mouse gut tissues, as well as the mesentery.15 While all have been shown to be effective in tissue clearing, Ce3D is rapid and requires five days, while some others entail longer processing times averaging 15 - 20 days. Our protocol enables high-resolution imaging in five days after fixation and is optimized to preserve small intestinal epithelial crypt-villus morphology and endogenous fluorescence. Our technique includes precise microdissection of intestinal tissue to enable spatial orientation of villi along radial or longitudinal axes tailored to specific biological inquiries. It also incorporates a custom-designed tissue imaging chamber, assembled from standard microscope slides and coverslips using liquid silicone and adhesive, that provides distinct advantages. Our imaging chamber accommodates both inverted and upright microscope configurations. We have leveraged this approach to enable a rapid and efficient method for high resolution 3D imaging of intestinal epithelial cells along the crypt-villus axis to fate map stem cells and their progeny.
Holding chambers to mount tissues can be made prior to tissue harvesting:
1% Phytagel/HD1.46 cylinder holding chamber.
-
1.
Using a cyanoacrylate adhesive (Superglue) stack 3 micro cover glasses (cover slips) (22 × 22 mm) on top of each other. Repeat the same for another stack of 3 micro cover slips (22 × 22 mm).
-
2.
Stick the first stack of 3 micro cover slips on the left-hand side of the Superfrost Plus Microscope slide using a cyanoacrylate adhesive (Superglue) and the other stack of 3 micro cover slips on the right-hand side, leaving a rectangular space in the middle.
-
3.
Using a 20 mL syringe and a 20G plastic needle, draw up 10 mL of liquid silicone and seal the 4 corners of the rectangular space.
Institutional permissions
All animal studies must be approved by the Institutional Animal Care and Use Committee (IACUC) and conducted in accordance with IACUC guidelines. The selection of mouse strain will depend on the specific experiment; to demonstrate this protocol, we employed lineage tracing of Fgfbp1+ cells in Fgfbp1-CreERT2; Rosa26-tdTomato mice aged 8–12 weeks.
Preparation of solutions required for fixing the intestine after harvesting
Timing: 15 min
-
4.Preparation of 2 mL 4% formaldehyde per sample.
-
a.Add 0.25 mL of “32% Paraformaldehyde”(EMS) to 1.75 mL of cold phosphate buffered saline (PBS).
-
a.
CRITICAL: Formaldehyde is listed as a Schedule 2, 6 and 8 hazardous classification and should be opened inside a fume hood. 4% formaldehyde should be prepared fresh, ideally an hour before harvesting the intestine.
-
5.Preparation of 2 mL of 30% sucrose per sample.
-
a.Dissolve 0.6 g of sucrose in 2 mL of 1X PBS and mix well.
-
b.Can be stored at 4°C for up to 24 h.
-
a.
Preparation of solutions required for optical clearing and 3D reconstruction
Timing: 2 h
-
6.Preparation of 0.1% PBS-Tween (PBST).
-
a.Mix 100 mL of 10X PBS, 1 mL of Tween-20, and 900 mL of double distilled (dd) H2O.
-
b.Can be stored at 20–25°C for up to 6 months.
-
a.
-
7.Preparation of 100 mL of 10% sodium azide (SAZ) stock solution.
-
a.Dissolve 10 g of SAZ in 100 mL of ddH2O and stir well with a magnetic stir bar.
-
b.Can be stored at 20–25°C for up to 4 months.
-
a.
CRITICAL: SAZ powder must only be handled inside a chemical fume hood while wearing a protective N95 mask. For the steps that follow, the 10% SAZ prepared will be used for preparing phosphate buffer (PB/SAZ), Histodenz, and 1% phytagel solution. Preparation of these solutions should also be handled and prepared in the hood.
-
8.Preparation of 10 mL of 1 M sodium hydroxide (NaOH).
-
a.Dissolve 0.5 g of NaOH powder in 10 mL of ddH2O and mix well.
-
b.Can be stored at 20–22°C for up to 4 months.
-
a.
-
9.Preparation of 150 mL delipidation buffer (8% sodium dodecyl sulfate [SDS]).
-
a.Add 50 ml of 10X PBS to 450 ml of ddH2O to make 500 mL of 1X PBS.
-
b.Dissolve 12 g of SDS in 130 mL of 1X PBS.
-
c.Stir well with a magnetic stir bar at 20–25°C.
-
d.Can be stored at 20–25°C for up to one month.
-
a.
CRITICAL: Ensure that pH is maintained at 9.25 with 1 M NaOH before use. Typically 40-50 μL of NaOH is required for adjusting the pH. A protective mask should be worn while handling SDS powder.
-
10.Preparation of 100 mL of 1 M sodium phosphate monobasic stock solution.
-
a.Add 15.6 g of sodium phosphate monobasic in 80 mL of ddH2O and stir well with a magnetic stirrer.
-
b.Add approximately 18 mL of ddH2O to make it up to exactly 100 mL.
-
c.Can be stored at 20°C–25°C for up to 6 months.
-
a.
-
11.Preparation of 100 mL of 0.5 M sodium phosphate dibasic stock solution.
-
a.Add 7.1 g of sodium phosphate dibasic in 80 mL of ddH2O and stir well with a magnetic stirrer until dissolved.
-
b.Add approximately 18 mL of ddH2O to make it up to exactly 100 mL.
-
c.Can be stored at 20°C–25°C for up to 6 months.
-
a.
-
12.
Preparation of 200 mL phosphate buffer with SAZ (PB/SAZ).
Note: See materials and equipment section for full recipe.
-
13.Preparation of 50 mL matching refractive index solution (HD1.46).
-
a.Dissolve 40 g of Histodenz in 25 mL of PB/SAZ.
-
b.Mix for 2 – 16 h at 20°C–25°C until complete dissolution.
-
c.Upon complete dissolution, bring volume up to 50 mL with PB/SAZ.
-
d.Can be stored at 4°C for up to 4 months.
-
a.
CRITICAL: Reagents with SAZ must only be handled inside a chemical fume hood while wearing a protective N95 mask. HD1.46 should be prepared in the fume hood.
-
14.Preparation of 10 mL 1% Phytagel solution.
-
a.Mix 0.1 g of Phytagel in 10 mL PB/SAZ on a hot plate set to 40°C with a magnetic stir bar.
-
b.For any remaining clumps, hot plate temperature can be increased to 55°C with stir bar to break up the clumps.
-
c.The remaining clumps can be further dissolved by heating solution in a microwave in 15 s intervals.
-
a.
CRITICAL: 1% Phytagel solution solidifies very quickly at 20°C–25°C and should immediately be transferred to a hot plate that is set at 40°C with a magnetic stir bar. There should be no bubbles or solids, but a homogenous solution.
CRITICAL: Reagents with SAZ must only be handled inside a chemical fume hood while wearing a protective N95 mask. 1% Phytagel should be prepared in the fume hood.
Note: Due to rapid solidification of 1% Phytagel, make this solution right before mounting tissues.
-
15.Preparation of 5 mL Phytagel/HD1.46 solution (1:1).
-
a.To prepare the solution, start by placing 5 mL of HD1.46 in a 45°C dry oven for at least 30 min to pre-warm it.
-
b.Next, in a 10 mL beaker placed on a hot plate set to 65°C, add 2.5 mL of the pre-warmed HD1.46 and begin stirring.
-
c.While stirring, gradually and continuously add 2.5 mL of a 1% Phytagel solution drop by drop. Increase stir speeds until the mixture achieves a uniform consistency.
-
a.
Note: Due to rapid solidification of the 1% Phytagel/HD1.46, make this solution right before mounting tissues.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Phosphate-buffered saline 10X | Sigma-Aldrich | Cat#P7059 |
| Phosphate-buffered saline 1X | Corning | Cat#21-040-CM |
| Paraformaldehyde 32% solution, EM grade | Electron Microscopy Sciences | Cat#15714-S |
| Sodium dodecyl sulfate | Sigma-Aldrich | Cat#L3771-100G |
| Sodium azide | Sigma-Aldrich | Cat#S2002 |
| DAPI | Roche | Cat#10236276001 |
| Sodium phosphate monobasic dihydrate | Sigma-Aldrich | Cat#71505Ā |
| Sodium phosphate dibasic | Sigma-Aldrich | Cat#S9763 |
| Histodenz | Sigma-Aldrich | Cat#D2158-100G |
| Phytagel | Sigma-Aldrich | Cat#P8169-250G |
| Sodium hydroxide pellets | Fisherbrand | Cat#S318-500 |
| Triton X-100 | Sigma-Aldrich | Cat#X100-100ML |
| Other | ||
| BD PrecisionGlide hypodermic needles – 20G × 1.5 inches | Fisher Scientific | Cat#305176 |
| Dissecting microscope | Nikon | SMZ1500 |
| 20 mL syringe | Fisherbrand | Cat#14955460 |
| Stir/hot plate | Fisherbrand Isotemp | Cat#SP88857200 |
| Magnetic stir bar | Fisherbrand | Cat#14-512-131 |
| Liquid silicone | General Electric | GE281 |
| Superfrost Plus microscope slides | Fisherbrand | Cat#1255015 |
| Micro cover glass (cover slip) | VWR | Cat#48366-067 |
| Micro cover glass (cover slip) | Fisherbrand | Cat#12541033 |
| Cyanoacrylate adhesive (Super Glue) | Gorilla | Cat#105796 |
| GenTeal Tears gel drop | Alcon | B0933LZVC9 |
| Experimental models: Organisms/strains | ||
| Fgfbp1-CreERT2; Rosa26-tdTomato mice males/females, 8–12 weeks | ||
| Software | ||
| LAS X | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
Materials and equipment
PBST
| Reagent | Final concentration | Amount |
|---|---|---|
| 10X PBS | 1X | 100 mL |
| Tween-20 | 0.1% | 1 mL |
| ddH2O | N/A | 900 mL |
| Total | N/A | 1000 mL |
Store at 20°C–25°C for up to 6 months.
PB/SAZ
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium azide | 0.01% | 200 μL |
| Sodium Phosphate Monobasic | 5.6 mM | 1.12 mL |
| Sodium Phosphate Dibasic | 14.4 mM | 5.76 mL |
| 1X PBS | 1X | 193 mL |
| Total | N/A | 200 mL |
Store at 20°C–25°C for up to 4 months.
HD1.46
| Reagent | Final concentration | Amount |
|---|---|---|
| PB/SAZ | 25 mL | |
| Histodenz | 40 g | |
| Total | N/A | 25 mL |
Bring volume up to 50 mL after dissolution of 40 g Histodenz. Store at 4°C for up to 4 months.
1% phytagel
| Reagent | Final concentration | Amount |
|---|---|---|
| PB/SAZ | 10 mL | |
| Phytagel | 0.1 g | |
| Total | N/A | 10 mL |
Store at 65°C on a hot plate with a magnetic stirrer for immediate use.
1% Phytagel/HD1.46 (1:1)
| Reagent | Final concentration | Amount |
|---|---|---|
| Histodenz 1.46 (HD1.46) | 50% | 2.5 mL |
| Phytagel 1% | 50% | 2.5 mL |
| Total | N/A | 5 mL |
Store at 65°C on a hot plate with a magnetic stirrer for immediate use.
Step-by-step method details
Harvesting of intestinal tissues
Timing: 20 min
This section details the approach for intestinal tissue harvesting, tissue clearing, fluorescent labeling, and 3D reconstruction.
-
1.Prepare sterile surgical instruments for harvesting intestinal tissues in addition to:
-
a.Glass beaker filled with 50 mL 1X PBS.
-
b.20 mL syringe attached with a 20-gauge needle.
-
c.1.2 mL of freshly prepared 4% formaldehyde solution in a 1.5 mL microcentrifuge tube.
-
a.
-
2.
Euthanize the mouse according to your institutional regulations.
-
3.
Spray down the abdomen with 70% ethanol and fix limbs down with needles.
-
4.
Cut open the abdominal cavity longitudinally and separately harvest the small intestine and colon.
-
5.
Flush out the luminal contents of the small intestine and colon with 1X PBS using 20 mL syringe and the 20-gauge needle.
Note: Mesenteric fat removal from the exterior of the intestines is recommended.
Fixation and water dispensation by hypertonic sucrose solution
Timing: 2 days
-
6.
Cut desired regions (duodenum, jejunum, ileum) into approximately 2 cm pieces.
-
7.Pass a needle through each of the 2 cm pieces to maintain tissue shape,
-
a.Place needle into a 1.2 mL of 4% formaldehyde in a 1.5 mL microcentrifuge tube.
-
b.Place the tubes in a microcentrifuge rack, cover with aluminum foil.
-
c.Place on ice-packs in the fume hood.
-
d.Remove the needles after 2 h until the intestine holds shape.
-
e.Close the microcentrifuge cap and transfer samples to 4°C.
-
a.
-
8.
Transfer tissue to a hypertonic 30% sucrose solution for 16-18 h to displace water molecules out of cells and prevent tissue shrinkage.
Note: 4% formaldehyde should be prepared with cold PBS and kept on ice.
Micro-dissection of fixed intestinal tissue
Timing: 1 h
-
9.
Prepare a Petri dish with fresh cold 1X PBS to prevent tissue from drying out.
-
10.
With fine scissors, cut tissue sections longitudinally, opening the intestine to lay flat with villi facing upward.
-
11.
Using a blade, micro-dissect consecutive strips of approximately 2 mm x 1 mm tissue under a dissection microscope.
CRITICAL: Ensure tissues pieces are uniform and 2 mm x 1 mm in size as this will ensure even clearing and refractive index matching across the tissue.
Note: For tissue samples exceeding 2 mm × 1 mm, we recommend extending both the clearing and refractive index matching durations. When mounting larger samples, a Cellvis 35 mm glass-bottom dish with a 20 mm microwell and #1 cover glass is suggested as the 1% Phytagel/HD1.46 chamber might be too small.
Delipidation of samples (tissue clearing) and wash
Timing: 2 days
In addition to proteins like collagen, lipids are another cellular component that exhibit a high refractive index (RI) in most biological tissues. Lipid content is particularly high in adipose tissue, making up over 70%, and around 20% in white matter and tongue tissue. In most soft tissues, lipids comprise about 10% of the tissue mass. The high RI of lipids contributes to light scattering within the tissue and also acts as a barrier to the penetration of external substances, as lipids form the primary component of cell membranes. Therefore, removing lipids is crucial for achieving thorough tissue clearing and allowing external molecules to permeate.
-
12.
Transfer tissues to 15 mL centrifuge tube with 10 mL delipidation buffer (8% SDS).
-
13.
Incubate in a rocking chamber at 42°C for 24 h.
CRITICAL: The delipidation buffer volume should be more than 1000X the sample size (e.g., intestinal tissue sections are approximately the size of a 10 μL liquid drop, a minimum of 10 mL buffer should be used).
-
14.
For a one-time wash, aspirate all delipidation buffer.
-
15.
Add 12 mL of 1X PBS to the tube.
-
16.
Incubate in a 42°C oven rocker for 24 h.
CRITICAL: The 1X PBS wash buffer should also be more than 1000X the sample size.
Nuclear staining and antibody
-
17.
Prepare a 1:500 dilution of DAPI in 12 mL of 1X PBST.
-
18.
Add 5 mL of 1X PBST to a single well of a 12-well plate.
-
19.
Transfer tissue sample from 1X PBS tube to the DAPI diluted PBST.
-
20.
Incubate on rocker at 20–25°C for 24 h.
CRITICAL: Gently separate tissue samples using forceps if they become stuck together.
-
21.
Wash samples 3 times with 1X PBST for 5 min each.
Note: If using primary and secondary antibodies, make sure to block with an appropriate blocking agent for your antibodies. Incubate tissues in a 1:100 dilution of primary antibody for 24 h, and incubate tissues in a 1:100 dilution of secondary antibody for 24 h. Wash for 10 min 3 times with 1X PBST after antibody staining steps.
Refractive index match
Timing: 1 day
The sample embedding medium surrounds the intestinal tissue during the mounting process. In biological samples, the RI of the medium is influenced by both the embedding medium and the glass cover slip, which typically has an RI between 1.47 and 1.52. To minimize refraction and achieve greater imaging depth, the embedding medium should have an RI similar to the lens immersion medium. Maintaining a uniform RI across the sample helps reduce diffraction. However, the RI can vary depending on the specific sample and cell type. Therefore, to minimize refraction and improve imaging quality, it is recommended to select an embedding medium that closely matches the RI of the sample. For the intestinal tissue 3D reconstruction and imaging, we have identified Histodenz 1.46 (HD1.46) as the best RI matching solution because of its ease of utility. Placing the sample in Histodenz for 24 h at 20–25°C allows the tissue to equilibrate so that its internal RI closely matches the medium. This reduces scattering when imaging the tissue.
-
22.
Transfer samples to a 2 mL microcentrifuge tube with 1.75 mL RI HD1.46 solution.
-
23.
Incubate on rocker at 20°C–25°C for 24 h. The sample is now ready for imaging.
-
24.
The samples can be kept at 4°C for longer-term storage for up to a month.
Preparing the stage
Timing: 30 min
-
25.
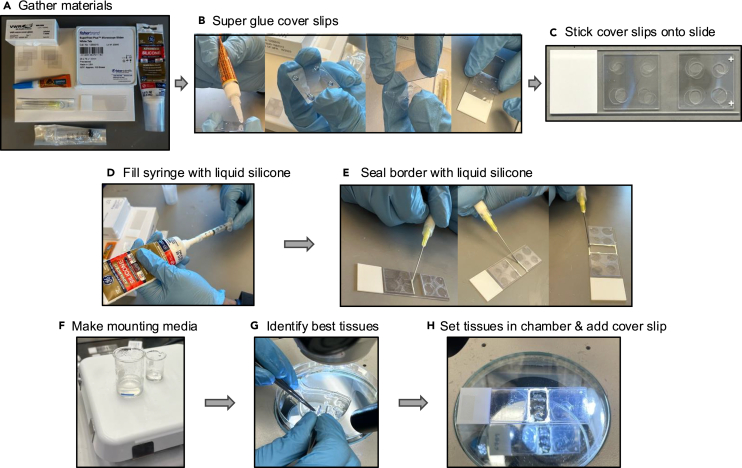
Gather all the materials required for stage preparation including micro cover slips, Superfrost Plus microscope slides, Superglue, liquid silicone, 20G needles, and 5 mL syringe (Figure 1A).
-
26.
Superglue two stacks of three 22 × 22 mm cover slips together (Figure 1B).
-
27.
Superglue the cover slip stacks on the left- and right-most part of a microscope slide, creating a 1 × 2 cm rectangular well in the middle of the slide (Figure 1C).
-
28.
Use liquid silicone to seal all four edges of the rectangular well in the middle of the slide (Figure 1D).
-
29.
Leave the stage to dry for 15-20 min (Figure 1E).
Figure 1.
Stage preparation and mounting of optically cleared intestinal tissues
(A) Gather materials including Superfrost Plus microscope slides, 22 × 22 mm micro cover slips, Superglue, liquid silicone, 20G needles, and 5 mL syringe.
(B) Assemble a stack of 3 cover slips with Superglue in between each cover slip. To assemble, take the first cover slip and apply Superglue on each corner and place second cover slip carefully on top. Apply Superglue on each corner of the second cover slip then carefully place third cover slip on top. Adhere the stack of glued cover slips onto a microscope slide.
(C) Repeat the assembly in B to make a second stack of three cover slips. Then Superglue the second stack on the other side of the microscope slide.
(D) Fill the 5 mL syringe with 2 mL liquid silicone.
(E) Attach a 20G needle to the syringe and carefully seal the borders in the center of the slide to create a chamber.
(F) Warm the HD1.46 and prepare the 1% phytagel. Then prepare the 1:1 HD1.46 and 1% phytagel solution, which will serve as the mounting solution.
(G and H) Identify the best cleared tissues and place 3 of them on the slide within the chamber. Add 100 μL of the mounting solution, immediately add a cover slip, and proceed to imaging or store in 4°C.
Making 1% Phytagel/HD1.46 mounting media
Timing: 15 min
-
30.
Warm 10 mL of HD1.46 at 45°C.
-
31.
On a 65°C hot plate, add 2.5 mL warm HD1.46 to a 10 mL beaker.
-
32.
Slowly add 2.5 mL of the 1% Phytagel solution, mixing with a magnetic bar until homogenous (Figure 1F).
Mounting samples onto the stage
Timing: 30 min
-
33.
Pre-warm the 2 mL microcentrifuge tube with tissue samples in Histodenz for 10 min at 45°C.
-
34.
identify best tissues in morphology and integrity and carefully place them in the rectangular well on the slide with villi transversely laying on the slide (Figure 1G).
-
35.
Using 200 μL pipette, add 100 μL of 1% Phytagel/HD1.46.
CRITICAL: Tissues may move upon addition of mounting media, quickly and gently adjust using forceps.
-
36.
Immediately cover the sample by placing the 24 x 50 mm microscope cover slip on top of the stacked cover slips and seal all the 4 edges with nail polish (Figure 1H).
Note: Due to the high density and viscous nature of 1% Phytagel/HD1.46, air bubbles seldom occur. However, if any do appear, carefully direct them to the chamber edge with small forceps and then break them.
-
37.
Immediately transfer slides to 4°C for 10 min.
-
38.
Use nail polish to seal all four edges of the cover slip.
Microscopy and image processing
-
39.
Power on the Stellaris laser scanning confocal microscope and launch the LAS X imaging acquisition software.
-
40.
Choose the 25x water immersion objective lens, apply a small drop of water-based solvent (Genteal tears gel) to the lens, position the sample chamber on the stage, and bring the sample into focus.
-
41.
Activate the fluorescence settings and locate your tissue using the DAPI channel.
-
42.
After locating the tissue, switch to confocal scanning mode and select the 568-nm laser to image the tdTomato labeled cells.
-
43.
Adjust the laser power to achieve a clear fluorescent signal for visualizing tdTomato-labeled cells, while avoiding excessive power to minimize photobleaching. Typically, the laser power is set to 2–6%, with the photomultiplier tube detector gain at 50 and the pinhole size adjusted to around 60 μm.
-
44.
To identify the central plane of the intestinal tissue, locate the bottom and top positions of the tissue by adjusting the focus.
-
45.
Optional: perform a z-scan to generate a stack of images for 3D reconstruction of the tissue, allowing visualization of different layers. Using z-steps of 5–9 μm is typically sufficient to determine the central plane of the tissue.
-
46.
Enable the 3D setting in the LAS X software and record videos of the tissue plane for a minimum of 1000 frames.
Note: Our images were acquired on an inverted Leica Stellaris 8 confocal microscope, run by LAS X software version 4.6.1. 27508, using a HC FLUOTAR L VISIR 25X/0.95 NA water immersion lens with Genteal tears gel as immersion fluid. The system includes a tunable 3rd generation WLL laser with HyD S, X, and R detectors. Unless noted, DAPI fluorescence was excited with a 405 nm DMOD laser set to 0.49% and collected on a HyD X detector with gain of 10 in intensity mode, range: 476-564 nm. The tdTomato was excited by the 554 nm WWL position set to 2.78%, with a HyD S detector gain at 35 in analog mode, range: 585-652 nm. The pinhole was 1 AU (55.9 μm), Z step size 0.567 μm, and X-Y pixel size was 0.606 μm, with dwell time of 1.575 μseconds. One hundred and one 2-channel z-sections were acquired by unidirectional scan of 1024 × 1024 pixels at 400 Hz scan speed for each image.
Note: This frame count, independent of the frame rate, ensures adequate data for analyzing lag times using LAS X. Capture more than 20 videos from 20 different samples of varying sizes to ensure robust data collection.
Expected outcomes
This protocol outlines a comprehensive step-by-step guide for intestinal tissue harvesting, optical clearing, and preparation for imaging and 3D reconstruction. Traditional approaches for lineage tracing in the small intestine often rely on detection of fluorescently labeled or immunofluorescence staining of 2D tissue samples, which limit accurate visualization of the 3D architecture of the crypt-villus axis. This makes it challenging to precisely quantify and localize intestinal epithelial cells within their spatial context. Here, we present a versatile optical clearing and imaging protocol that is less labor-intensive and significantly faster than most existing methods, requiring only five days of clearing, totaling seven days including fixation. Our approach provides a balance between speed, ease of use, while preserving of intestinal crypt-villus morphological integrity and endogenous fluorescence. We demonstrate this technique within the context of lineage tracing in the Fgfbp1-CreERT2; Rosa26-tdTomato mouse intestine. Following tamoxifen administration, the intestinal tissue was harvested, delipidated, stained with the nuclear dye DAPI, RI-matched, and mounted (Figure 2, Methods video S1). This entire process was completed within seven days, highlighting the rapid and efficient protocol that enables precision localization of endogenous fluorescence within the crypt-villus architecture of the small intestine.
Figure 2.
Whole mount staining of cleared intestinal tissues
(A) Visualization of the mouse intestinal tissue with nuclear stain DAPI.
(B) Visualization of the mouse intestinal tissue with Fgfbp1+ cells labeled with tdTomato.
(C) Example 3D projection images of a whole-mount staining of intestinal tissues showing endogenous tdTomato reporter labeling Fgfbp1+ cells with DAPI. Scale bar = 100 μm. Images were taken with a Leica STELLARIS 8 Confocal Microscope.
3D reconstruction of whole-mount intestinal tissues stained with DAPI showing endogenous tdTomato labeling of Fgfbp1+ cells and their progeny. Scale bar = 100 μm.
Limitations
We have optimized this protocol for intestinal tissue samples up to 2 mm × 1 mm in size. While the tissue appears slightly less transparent towards the center at these dimensions, it remains sufficiently clear for whole-tissue imaging. We anticipate that clearing thicker tissue samples may pose increasing challenges; however, this protocol has not been tested under such conditions. Since tissue clarity improves with extended incubation in both the initial delipidation step and the Histodenz RI matching solution, we expect that longer clearing and RI matching times could enable effective clearing of much thicker specimens, provided sufficient time is allowed.
We anticipate that an 8% SDS solution at pH 9.25 combined with Histodenz 1.46 will effectively clear most 4% formaldehyde-fixed intestinal tissues. This method has shown satisfactory results across different regions of the small intestine as well as the colon. However, other tissue types may require modifications to the clearing protocol, such as incorporating electrophoresis, adjusting incubation times, or using alternative RI matching reagents. These variations may depend on factors such as species, genetic background, age, disease state, or experimental conditions. To ensure optimal results, it is essential to empirically determine the specific clearing conditions for each tissue type before initiating an experiment.
This protocol provides a detailed guide for optical clearing and 3D reconstruction of intestinal tissues fixed in 4% formaldehyde, and the imaging performed using the STELLARIS 8 confocal microscope which features a unique 3D machine learning algorithm that enhances reconstruction accuracy. However, certain antibodies may require alternative fixation conditions to preserve appropriate signal detection. For tissues necessitating fixation methods other than 4% formaldehyde, further optimization of tissue preparation steps may be required. Additionally, users employing imaging systems other than STELLARIS 8 confocal microscope such as light sheet microscopes should optimize their imaging protocols to achieve comparable results.
Troubleshooting
Problem 1: Microbial contamination in tissue processing
During tissue processing, which spans over seven days with prolonged incubation at 42°C, conditions can become favorable for the growth of bacteria or fungi. Signs of microbial contamination may include cloudiness in the buffer solution surrounding the tissue or the appearance of whitish-yellow growths on the tissue itself. Additionally, the presence of an unpleasant, putrid odor can serve as a strong indication of contamination.
Potential solution
Contaminated tissue should be discarded to avoid compromising results. To minimize the risk of contamination, it is essential to filter the 8% SDS solution through a 0.2 μm filter after adjusting the pH. Additionally, all solutions used for tissue incubation, treatment, and storage (excluding the initial clearing solution) must contain a high concentration of sodium azide (0.01%). Sodium azide is highly toxic; therefore, strict precautions should be taken to prevent skin contact or accidental ingestion.
Problem 2: Opacity in the center of intestinal tissue after clearing
The central portion of the intestinal tissue often appears more opaque than the edges following the clearing process. This is because the clearing process primarily depends on the passive or active diffusion of lipids and other substances through the tissue. As a result, smaller regions like the edges clear more efficiently, while the center, especially in larger tissue samples exceeding 2 mm × 1 cm, can be more challenging to clear.
Potential solution
Several factors contribute to the persistence of opacity in the center of intestinal tissue. A primary cause is incomplete diffusion of micelles during the clearing process, often resulting from insufficient rotation of the tissue in the 42°C incubator. One common issue is overfilling the 15 mL tube with 15 mL of clearing solution, which causes the tissue to float and inhibits proper agitation. To address this, we recommend using only 14 mL of clearing solution in a 15 mL tube to ensure consistent and effective tissue agitation throughout the process.
Another important factor influencing tissue opacity is sample size. Intestinal tissues exceeding 2 mm × 1 mm typically require extended processing times, including more than 24 h of SDS-based clearing and over 24 h of refractive index matching with Histodenz to achieve uniform transparency. For mounting and imaging larger samples, the standard 1% Phytagel/HD1.46 chamber may not be sufficient. In these cases, we recommend using a Cellvis 35 mm glass-bottom dish with a 20 mm microwell and #1 cover glass.
Problem 3: Leakage of mounting solution and tissue displacement
During sample preparation, mounting solution (1% Phytagel/HD1.46) may occasionally leak onto the edges and into the cover slips, particularly after placing the cover slip on the slide. After positioning the intestinal tissue at the center using forceps and adding 250 μL of mounting media, the solution can sometimes spread unevenly. This unintended dissipation can displace the tissue, resulting in a loss of its intended orientation and alignment.
Potential solution
This issue is often caused by improper sealing of the rectangular area between cover slips with liquid silicone. To correct this, carefully remove the glass slide and use forceps to gently detach the tissue. Transfer the tissue to a fresh tube containing HD1.46 to preserve its condition. Clean the slide thoroughly with 1X PBS and allow it to air dry for 5 min. Reseal the edges with liquid silicone, ensuring a secure seal to prevent leakage in subsequent preparations.
Problem 4: Air bubbles in the mounting solution (1% Phytagel/HD1.46)
While positioning the tissue during sample mounting, we apply the mounting solution 1% Phytagel/HD1.46. Occasionally, air bubbles may form within this solution, causing potential light scattering issues.
Potential solution
Due to the high density and viscosity of 1% Phytagel/HD1.46, air bubbles are uncommon. However, if bubbles do form, gently move them to the edge of the chamber using small forceps and burst them.
Problem 5: Tissue entanglement after delipidation, PBS wash, and nuclear staining
Due to the rotating and rocking motions that tissues undergo during lipidation, washing, and DAPI staining, intestinal tissues can occasionally become entangled with each other.
Potential solution
Place the intestinal tissues into a petri dish containing 1X PBS to minimize entanglement. If tissues become stuck together during processing steps, gently separate them using fine-tipped forceps, taking care not to damage or tear the samples.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kelley Yan (ky2004@cumc.columbia.edu).
Technical contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the technical contact, Joel George (jjg2216@cumc.columbia.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate or analyze any code.
Acknowledgments
This work was funded by NIHDP2DK128801, NIHR01AG067014, NIHR01DK142438, BWF CAMS, the Gerstner Foundation, the Irma T. Hirschl Trust, and the Irving Scholars Award to K.S.Y. and NIH U01DK103155, as part of the Intestinal Stem Cell Consortium (NIDDK/NIAID), P30CA013696, P30DK132710, and S10OD032447. C.C. was supported by an NYSTEM predoctoral award, E.W. by NSFGRF #2036197, J.M. by NIH T32DK083256, L.C. and H.L. by the Berrie Foundation, and J.J.G. by NIH T32HL105323. Imaging was performed in the Columbia Medicine Microscopy Core.
Author contributions
J.J.G., E.W., S.H.B., and C.C. developed the clearing and refractive index matching protocol. R.T.S., J.W.M., and H.-W.S. provided technical guidance. F.M., H.L., J.M., and L.C. provided technical and experimental support. J.J.G., E.W., S.H.B., and K.S.Y. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2025.103841.
Contributor Information
Joel Johnson George, Email: jjg2216@cumc.columbia.edu.
Kelley S. Yan, Email: ky2004@cumc.columbia.edu.
References
- 1.Capdevila C., Miller J., Cheng L., Kornberg A., George J.J., Lee H., Botella T., Moon C.S., Murray J.W., Lam S., et al. Time-resolved fate mapping identifies the intestinal upper crypt zone as an origin of Lgr5+ crypt base columnar cells. Cell. 2024;187:3039–3055.e14. doi: 10.1016/j.cell.2024.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 3.Tian H., Biehs B., Warming S., Leong K.G., Rangell L., Klein O.D., de Sauvage F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan K.S., Janda C.Y., Chang J., Zheng G.X.Y., Larkin K.A., Luca V.C., Chia L.A., Mah A.T., Han A., Terry J.M., et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature. 2017;545:238–242. doi: 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung K., Wallace J., Kim S.Y., Kalyanasundaram S., Andalman A.S., Davidson T.J., Mirzabekov J.J., Zalocusky K.A., Mattis J., Denisin A.K., et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Y.Y., Tang S.C. Optical clearing facilitates integrated 3D visualization of mouse ileal microstructure and vascular network with high definition. Microvasc. Res. 2010;80:512–521. doi: 10.1016/j.mvr.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Fu Y.Y., Lin C.W., Enikolopov G., Sibley E., Chiang A.S., Tang S.C. Microtome-free 3-dimensional confocal imaging method for visualization of mouse intestine with subcellular-level resolution. Gastroenterology. 2009;137:453–465. doi: 10.1053/j.gastro.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu Y.Y., Peng S.J., Lin H.Y., Pasricha P.J., Tang S.C. 3-D imaging and illustration of mouse intestinal neurovascular complex. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;304:G1–G11. doi: 10.1152/ajpgi.00209.2012. [DOI] [PubMed] [Google Scholar]
- 9.Ke M.T., Fujimoto S., Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 2013;16:1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- 10.Kaufman J.A., Castro M.J., Sandoval-Skeet N., Al-Nakkash L. Optical clearing of small intestine for three-dimensional visualization of cellular proliferation within crypts. J. Anat. 2018;232:152–157. doi: 10.1111/joa.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatoko T., Harada N., Tokumoto S., Yamane S., Ikeguchi-Ogura E., Kato T., Yasuda T., Tatsuoka H., Shimazu-Kuwahara S., Yabe D., et al. An analysis of intestinal morphology and incretin-producing cells using tissue optical clearing and 3-D imaging. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-22511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller C.E., Thompson R.P., Bigelow M.R., Gittinger G., Trusk T.C., Sedmera D. Confocal imaging of the embryonic heart: how deep? Microsc. Microanal. 2005;11:216–223. doi: 10.1017/S1431927605050464. [DOI] [PubMed] [Google Scholar]
- 13.Li W., Germain R.N., Gerner M.Y. Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (C(e)3D) Proc. Natl. Acad. Sci. USA. 2017;114:E7321–E7330. doi: 10.1073/pnas.1708981114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bossolani G.D.P., Pintelon I., Detrez J.D., Buckinx R., Thys S., Zanoni J.N., De Vos W.H., Timmermans J.P. Comparative analysis reveals Ce3D as optimal clearing method for in toto imaging of the mouse intestine. Neuro Gastroenterol. Motil. 2019;31 doi: 10.1111/nmo.13560. [DOI] [PubMed] [Google Scholar]
- 15.Neckel P.H., Mattheus U., Hirt B., Just L., Mack A.F. Large-scale tissue clearing (PACT): Technical evaluation and new perspectives in immunofluorescence, histology, and ultrastructure. Sci. Rep. 2016;6 doi: 10.1038/srep34331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3D reconstruction of whole-mount intestinal tissues stained with DAPI showing endogenous tdTomato labeling of Fgfbp1+ cells and their progeny. Scale bar = 100 μm.
Data Availability Statement
This study did not generate or analyze any code.