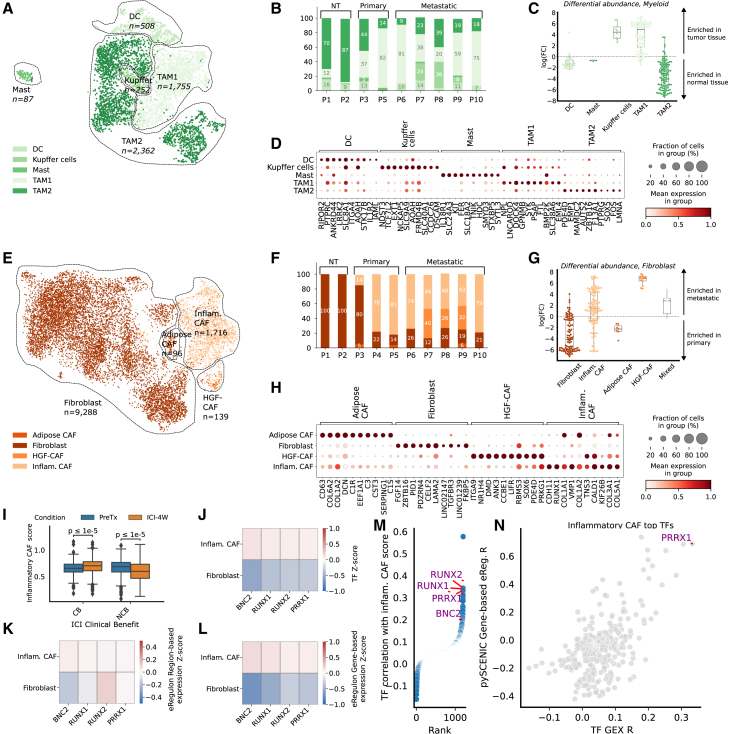
Figure 2.
The EAC TME contains several pro- and anti-inflammatory populations of macrophages and RUNX1/RUNX2/PRRX1/BNC2-regulated inflammatory cancer-associated fibroblasts enriched in metastatic samples
(A) Uniform manifold approximation and projection (UMAP) representation of the myeloid compartment in Harmony-corrected integrated transcriptomic data, with annotated subtypes indicated.
(B) Proportion of myeloid subtypes per patient.
(C) Distribution of Milo20 fold change scores between normal-adjacent and tumor samples for myeloid cells; Milo scores measure differential abundances of specific cell subtypes by assigning cells to overlapping neighborhoods in a k-nearest neighbor graph.
(D) Marker genes of annotated myeloid subtypes, with cells grouped by subtype and expression information provided.
(E) UMAP representation of the fibroblast compartment in Harmony-corrected integrated transcriptomic data, with annotated subtypes indicated.
(F) Proportion of fibroblast subtypes per patient.
(G) Distribution of Milo fold change scores between metastatic and primary tumor samples for fibroblast subtypes, with labeling and exclusion criteria similar to (C).
(H) Marker genes of annotated fibroblast subtypes, with cells grouped by subtype and expression information provided.
(I) Distribution of the inflammatory cancer-associated fibroblast (CAF) score in the stromal compartment of Carroll et al.’s9 cohort, stratified by response to immune checkpoint inhibitor (ICI) therapy: clinical benefit (CB) and no clinical benefit (NCB). The inflammatory CAF program is scored on the entire cohort. Paired measurements of patients were made before treatment (PreTx) and after a 4-week ICI treatment window (ICI-4W). The distribution of the inflammatory CAF score is compared among the CB and NCB groups across PreTx and ICI-4W time points. Significance testing is conducted using a Mann-Whitney test to assess differences between the CB and NCB groups.
(J–L) Results for SCENIC+-derived transcription factor (TF) candidates for inflammatory fibroblasts, with cells grouped by subtype and Z scores of TF expression (J), eRegulon gene-based expression (K), and eRegulon region-based expression (L) are shown.
(M) TF gene expression correlation with inflammatory CAF score in the external pan-cancer fibroblast validation cohort of Luo et al.,16 with candidate TFs identified with the SCENIC+ analysis highlighted.
(N) Correlation of all available TFs’ gene expression and SCENIC-estimated gene-based eRegulon score with the inflammatory CAF score in the pan-cancer fibroblast atlas.16 Only PRRX1’s eRegulon activity, but not BNC2 and RUNX1/2, was estimated using SCENIC.