Table 1.
Indicators for model predictions of Cα AMSDs for full protein set
| Model | Density* | 〈Δ〉† | Range of Δ | 〈ρ〉† | Range of ρ | |
|---|---|---|---|---|---|---|
| a | LDM | all/ref/fix | 0.89 ± 0.27 | 0.63–2.19 | 0.62 ± 0.09 | 0.41–0.80 |
| b | LDM | all/ref/scd | 0.86 ± 0.26 | 0.62–2.09 | 0.64 ± 0.09 | 0.43–0.81 |
| c | LDM | all/xtl/fix | 0.75 ± 0.12 | 0.52–1.21 | 0.67 ± 0.09 | 0.45–0.83 |
| d | LDM | all/xtl/scd | 0.72 ± 0.11 | 0.53–1.13 | 0.70 ± 0.09 | 0.49–0.85 |
| e | P-GNM | Cα/ref/fix | 1.08 ± 0.42 | 0.65–3.06 | 0.58 ± 0.17 | 0.05–0.84 |
| f | LDM | Cα/ref/fix | 1.02 ± 0.32 | 0.74–2.58 | 0.51 ± 0.11 | 0.20–0.70 |
| g | LDM | Cα/ref/scd | 0.97 ± 0.29 | 0.68–2.32 | 0.58 ± 0.08 | 0.42–0.75 |
In all calculations, the cutoff radius RC was set equal to the distance, R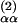 , of the second minimum in the Cα–Cα radial density.
, of the second minimum in the Cα–Cα radial density.
Contact density is based on nonhydrogen atoms (all) or Cα atoms (Cα) in reference molecule (ref) or entire crystal (xtl) and is calculated with fixed (fix) or self-consistently distributed (scd) atoms.
Mean value ± one standard deviation.
