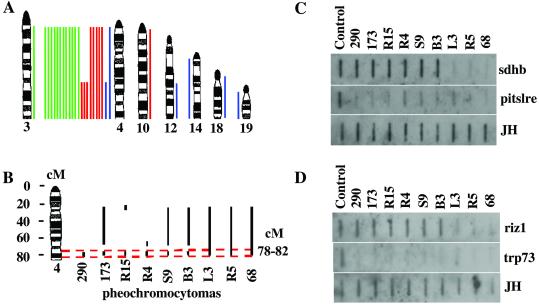
Figure 4.
Chromosomal alterations detected by conventional CGH and array-CGH in mouse pheochromocytomas. (A) Lines to the right of the ideograms indicate gains; lines to the left indicate losses. Pheochromocytomas from 4 Pten+/− mutant mice (blue), 8 Ink4a/Arf+/− Pten+/− (red), and 12 Ink4a/Arf−/− Pten+/− (green) mice were analyzed by conventional CGH. (B) Schematic illustration of deleted regions in chromosome 4 by array-CGH. Vertical bars indicate regions deleted. Horizontal dashed lines define the most commonly deleted region. (C and D) Slot blot analysis of mouse sdhd and pitslre, riz1 and trp73 genes. One microgram of normal or tumor genomic DNA was loaded in each well. Corresponding DNA fragments were radiolabeled as probes. A DNA fragment of mouse Ig JH gene was used to confirm the DNA quantity and quality.