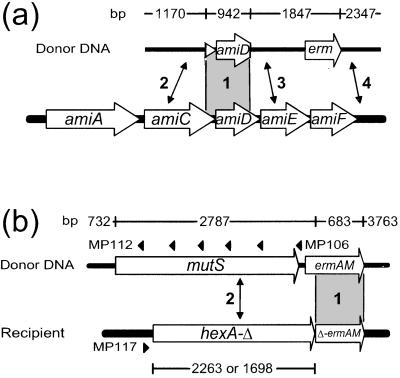
Figure 2.
Experimental systems to investigate InsDel formation and the capture of foreign DNA. Lengths of donor segments are indicated above the map. (a) The R800 wild-type recipient was transformed by using as donor purified PstI-linearized plasmid pR313 DNA. (b) Purified HindIII-linearized plasmid pCHS101 DNA was used to transform the TCHA2 or TCHA3 recipients. Diagnostic oligonucleotides used as PCR primers (MP117 combined with MP112 to MP116 or MP106) for the analysis of recombinants are indicated by black triangles (numbering of MP113 to MP116, located from left to right between MP112 and MP106, was omitted for clarity). The homologous anchor provided by the amiC–amiD or the ermAM segments, present in both donor and recipient DNA in a and b respectively, is shown in gray (region 1). While NJs formed in regions 2, 3, or 4 in a lead to InsDels (scored as MTXR transformants), only those formed in region 4 promote capture of the ermAM gene (EryR transformants). (b) NJs formed to the left of ermAM (region 2) restore a functional ermAM gene. The truncated hexA coding region is 2,263 bp (49.1% identity with mutS) or 1,698 bp long (46.9% identity with mutS) in strain TCHA2 and TCHA3, respectively (Materials and Methods).