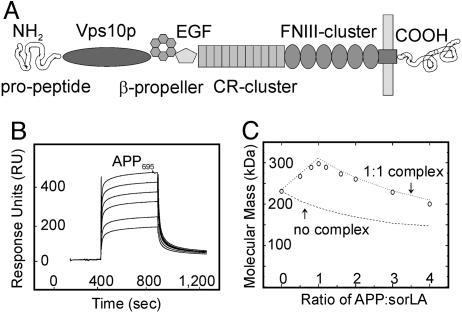
Fig. 1.
APP and sorLA interact in vitro.(A) The structural elements of sorLA are depicted, including (from amino to carboxyl terminus) the propeptide, vacuolar protein sorting 10 protein (Vps10p) domain, β-propeller, epidermal growth factor repeat (EGF), and clusters of complement-type repeats (CR) and fibronectin type III domains (FNIII). (B and C) The sorLA ectodomain used for surface plasmon resonance (SPR) (B) or molecular mass analysis (C) encompassed all elements amino-terminal to the membrane anchor. (B) SPR analysis of binding of a concentration series (0.1, 0.2, 0.5, 1, 2, and 5 μM) of APP695 to sorLA immobilized on the sensor chip (KD = 200 nM). (C) Molecular-weight plot of APP-sorLA complex formation by analytic ultracentrifugation. The sorLA concentration was kept at 0.1 μM, whereas the APP695 concentration varied to give the indicated APP:sorLA input ratios. Circles indicate the actual data points of the observed average molecular masses in the protein mixture at a given APP:sorLA ratio. Lines represent theoretical graphs to be expected for a perfect 1:1 complex (dotted) or no complex formation (dashed).