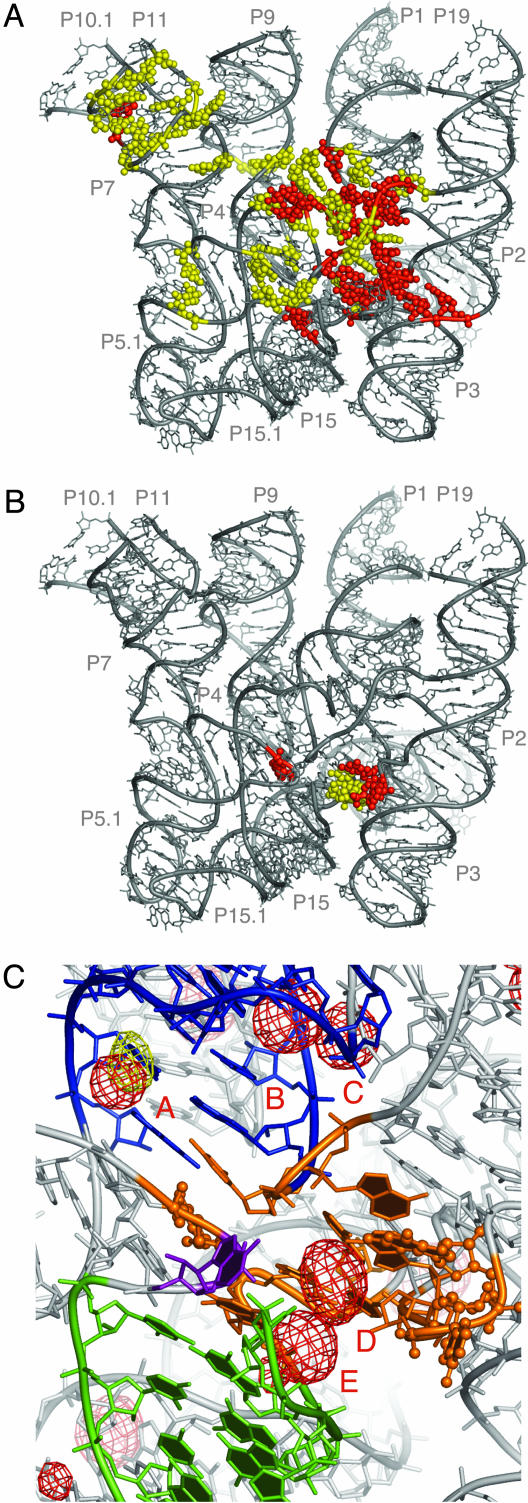
Fig. 4.
Base conservation in the bacterial RNase P RNA and identification of the chemically active site. (A) Distribution of the conserved nucleotides in the bacterial RNase P RNA. Nucleotides absolutely conserved in bacteria are highlighted in red, whereas those highly conserved (≥80%) are shown in yellow. (B) Nucleotides in bacterial RNase P RNA crosslinked by a photoagent at the 5′ end of product tRNA (9). Nucleotides crosslinked in both A- and B-types of RNase P RNA are colored in red, whereas nucleotides crosslinked only in the A-type RNase P RNA are colored in yellow. (C) Proposed chemically active site (orange). Balls and sticks, nucleotides crosslinked by a photoagent at the 5′ end of product tRNA in A- and B-types of RNase P RNA. Helix P4 is colored in blue, P15 in green, and nucleotide and A256 in purple. RNA-phased anomalous difference maps from Os(III) hexamine (red mesh) and Pb(II) (yellow mesh) contoured at 4.5 σ indicate potential metal-binding sites. The metal-bindng sites discussed in the text are labeled A through E. The orientation of the model is approximately the same as in Fig. 3B.