Figure 6.
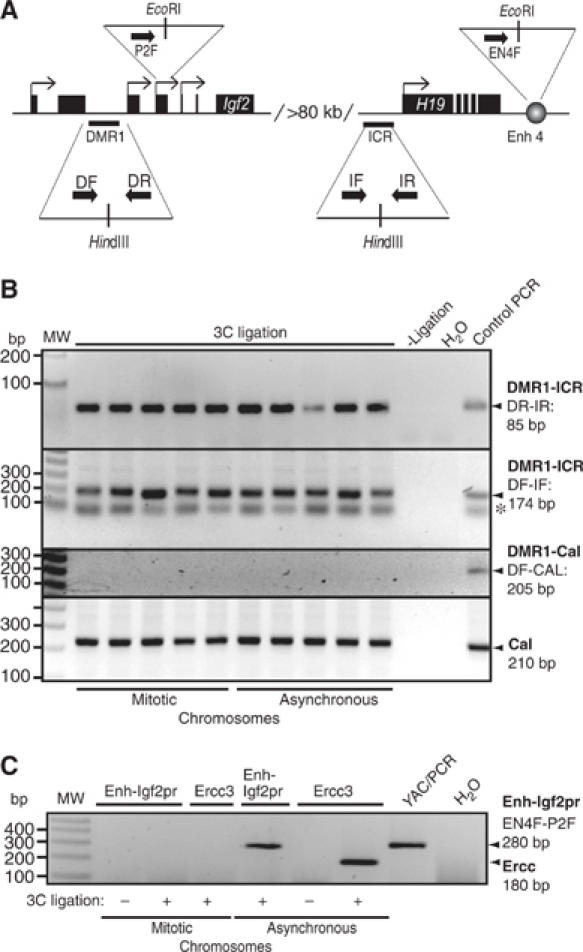
The Igf2 DMR1–H19 ICR interaction is an epigenetic mark. (A) Schematic representation of the primers used. The Igf2 DMR1 and H19 ICR as well as Igf2 promoter 2 and H19 enhancers, which are separated by more than 80 kb of intervening sequences, are indicated in the image. The primers used are DF and DR for Igf2 DMR1, IF and IR for H19 ICR, P2F for the Igf2 promoter 2 and EN4F for the enhancer 4 downstream of H19. (B) Long-distance ICR/DMR1 interaction is maintained in mitotic chromosomes. Chromatin interaction between the H19 ICR and Igf2 DMR1 was studied by 3C analysis using chromatin from NIH3T3 cells and two different primer combinations. PCR products from primer combinations DR plus IR, and DF plus IF were run on 2.5 and 1.5% agarose gel, respectively, with ethidium bromide staining. The last lane in both the gels shows the PCR products with template obtained after digestion and ligation of YAC (for Igf2 DMR1 and H19 ICR) or a control plasmid (containing the DMR1 and Calreticulin sequences, see below). The band in all the lanes labeled with an asterisk is a nonspecific band, which does not hybridize to a specific DMR1 probe (not shown). All individual PCR analyses of five independent 3C samples were performed at least twice. To account for random background interactions, we performed PCR using one primer each from Calreticulin (CalR) and Igf2 DMR1 as indicated in the panel. The control template for this analysis was generated by mixing, digesting and ligating CalR and Igf2 DMR1 HindIII amplicons in equimolar amounts. The CalR products were used for normalizing signals to account for crosslinking efficiency of mitotic/asynchronous chromosomes with previously used primers (Tolhuis et al, 2002). (C) Long-distance enhancer/promoter interaction is not maintained in mitosis. One of the samples (#3 in both mitotic and asynchronous chromosome preparations) presented in panel B was subjected to additional 3C analysis to test for unspecific interaction. The interactions within the Ercc3 locus (encompassing 14 kb) (Palstra et al, 2003) and between the H19 enhancer and Igf2 promoter 2 (encompassing 101 kb) (Tiwari et al, unpublished observation) were examined in mitotic and asynchronous chromosomes. In both cases, the interaction seen in asynchronous cells (Ercc and Enh-Igf2pr) is lost in mitotic chromatin. Signals were always dependent on the ligation step (3C ligation).
