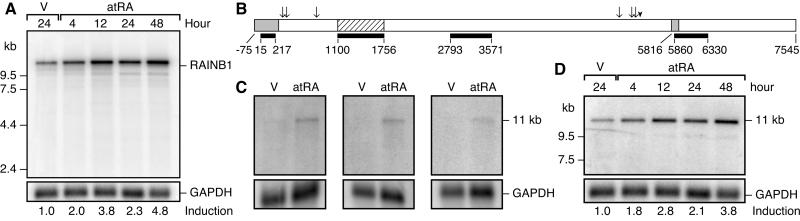
Figure 1.
Induction of RAINB1 by atRA. (A) Northern blot analysis of RAINB1 mRNA from SH-SY5Y cells treated with atRA (10−7 M) or 24 h with vehicle. (B) A schematic representation of the cDNA amplified from reverse transcribed mRNA and the cDNA probes used for the Northern blot analysis. The A of the ATG start site of the predicted ORF is designated nucleotide +1, and the sequence is numbered accordingly. The striped box represents the original 657-bp RAINB1 cDNA clone, and the black bars shown beneath represent the location of the cDNA probes used in Northern blotting studies. The gray boxes (the most 5′ prime region and site of 101-bp deletion starting at nucleotide 5816) and the arrowhead (site of 9-bp deletion) indicate the regions that can vary between the 12 related sequences from the patent database (AX034574 and AX009316–26). The arrows indicate the six nucleotide positions where the RAINB1 sequence differs from the AX009316 sequence. (C) mRNA from SH-SY5Y cells treated for 16 h with atRA or vehicle was probed with the original RAINB1 cDNA (nucleotides 1100–1756) (Left), the probe ranging from nucleotides 2793 to 3571 (Center), and that from nucleotides 5860 to 6330 (Right). (D) Northern blot analysis of the membrane shown in A with the probe comprising nucleotides 15–217. Hybridization with a radiolabeled glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probe (nucleotides 11–1053 within the ORF of the human cDNA) was used to correct for any differences in mRNA loaded when calculating fold-induction.